Abstract
Introduction
Oral epithelial dysplasia (OED) is a precancerous histopathological finding which is considered the most important prognostic indicator for determining the risk of malignant transformation into oral squamous cell carcinoma (OSCC). The gold standard for diagnosis and grading of OED is through histopathological examination, which is subject to inter- and intra-observer variability, impacting accurate diagnosis and prognosis. The aim of this review article is to examine the current advances in digital pathology for artificial intelligence (AI) applications used for OED diagnosis.
Materials and Methods
We included studies that used AI for diagnosis, grading, or prognosis of OED on histopathology images or intraoral clinical images. Studies utilizing imaging modalities other than routine light microscopy (e.g., scanning electron microscopy), or immunohistochemistry-stained histology slides, or immunofluorescence were excluded from the study. Studies not focusing on oral dysplasia grading and diagnosis, e.g., to discriminate OSCC from normal epithelial tissue were also excluded.
Results
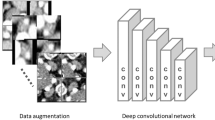
A total of 24 studies were included in this review. Nineteen studies utilized deep learning (DL) convolutional neural networks for histopathological OED analysis, and 4 used machine learning (ML) models. Studies were summarized by AI method, main study outcomes, predictive value for malignant transformation, strengths, and limitations.
Conclusion
ML/DL studies for OED grading and prediction of malignant transformation are emerging as promising adjunctive tools in the field of digital pathology. These adjunctive objective tools can ultimately aid the pathologist in more accurate diagnosis and prognosis prediction. However, further supportive studies that focus on generalization, explainable decisions, and prognosis prediction are needed.

Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
References
Woo S-B (2019) Oral epithelial dysplasia and premalignancy. Head Neck Pathol 13(3):423–439. https://doi.org/10.1007/s12105-019-01020-6
Wenig BM (2017) Squamous cell carcinoma of the upper aerodigestive tract: dysplasia and select variants. Mod Pathol 30:S112–S128. https://doi.org/10.1038/modpathol.2016.207
van der Waal I (2009) Potentially malignant disorders of the oral and oropharyngeal mucosa; terminology, classification and present concepts of management. Oral Oncol 45(4):317–323. https://doi.org/10.1016/j.oraloncology.2008.05.016
Warnakulasuriya S, Ariyawardana A (2016) Malignant transformation of oral leukoplakia: a systematic review of observational studies. J Oral Pathol Med 45(3):155–166. https://doi.org/10.1111/jop.12339
Khoury ZH, Sultan M, Sultan AS (2022) Oral epithelial dysplasia grading systems: a systematic review & meta-analysis. Int J Surg Pathol 30(5):499–511. https://doi.org/10.1177/10668969211070171
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444. https://doi.org/10.1038/nature14539
Fisher RB, Breckon TP, Dawson-Howe K et al (2013) Dictionary of computer vision and image processing. Wiley
Mahmood H, Shaban M, Indave B et al (2020) Use of artificial intelligence in diagnosis of head and neck precancerous and cancerous lesions: a systematic review. Oral Oncol 110:104885. https://doi.org/10.1016/j.oraloncology.2020.104885
Baik J, Ye Q, Zhang L et al (2014) Automated classification of oral premalignant lesions using image cytometry and random forests-based algorithms. Cell Oncol 37(3):193–202. https://doi.org/10.1007/s13402-014-0172-x
Haralick RM, Shapiro LG (1985) Image segmentation techniques. Comput Vis Graph Image Proc 29(1):100–132. https://doi.org/10.1016/S0734-189X(85)90153-7
Liu Y, Bilodeau E, Pollack B et al (2022) Automated detection of premalignant oral lesions on whole slide images using convolutional neural networks. Oral Oncol 134:106109. https://doi.org/10.1016/j.oraloncology.2022.106109
Shephard A, Azarmehr N, Bashir RMS et al (2022) A fully automated multi-scale pipeline for oral epithelial dysplasia grading and outcome prediction. Medical Imaging with Deep Learning
Zhang X, Gleber-Netto FO, Wang S et al (2023) Deep learning-based pathology image analysis predicts cancer progression risk in patients with oral leukoplakia. Cancer Med. https://doi.org/10.1002/cam4.5478
Nguyen P-T-H, Sakamoto K, Ikeda T (2022) Deep-learning application for identifying histological features of epithelial dysplasia of tongue. J Oral Maxillofac Surg Med Pathol 34(4):514–522. https://doi.org/10.1016/j.ajoms.2021.12.008
da Rocha K, Bermudez JCM, Rivero ERC et al (2022) A pathology-based machine learning method to assist in epithelial dysplasia diagnosis. Res Biomed Eng 38(3):989–1002. https://doi.org/10.1007/s42600-022-00234-y
Azarmehr N, Shephard A, Mahmood H et al (2021) Automated oral epithelial dysplasia grading using neural networks and feature analysis. Medical Imaging with Deep Learning
Taha AA, Hanbury A (2015) Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 15:29. https://doi.org/10.1186/s12880-015-0068-x
Ellis BG, Whitley CA, Triantafyllou A et al (2022) Prediction of malignant transformation in oral epithelial dysplasia using infrared absorbance spectra. PLoS ONE 17(3):e0266043. https://doi.org/10.1371/journal.pone.0266043
Ferrer-Sánchez A, Bagan J, Vila-Francés J et al (2022) Prediction of the risk of cancer and the grade of dysplasia in leukoplakia lesions using deep learning. Oral Oncol 132:105967. https://doi.org/10.1016/j.oraloncology.2022.105967
Ingham J, Smith CI, Ellis BG et al (2022) Prediction of malignant transformation in oral epithelial dysplasia using machine learning. Iop Scinotes 3(3):034001. https://doi.org/10.1088/2633-1357/ac95e2
da Rocha K, Bermudez JC, Rivero ER et al (2022) A pathology-based machine learning method to assist in epithelial dysplasia diagnosis. Res Biomed Eng 38(3):989–1002. https://doi.org/10.1007/s42600-022-00234-y
Camalan S, Mahmood H, Binol H et al (2021) Convolutional neural network-based clinical predictors of oral dysplasia: class activation map analysis of deep learning results. Cancers (Basel) 13(6). https://doi.org/10.3390/cancers13061291
Duran-Sierra E, Cheng S, Cuenca R et al (2021) Machine-learning assisted discrimination of precancerous and cancerous from healthy oral tissue based on multispectral autofluorescence lifetime imaging endoscopy. Cancers 13(19):4751. https://doi.org/10.3390/cancers13194751
James BL, Sunny SP, Heidari AE et al (2021) Validation of a point-of-care optical coherence tomography device with machine learning algorithm for detection of oral potentially malignant and malignant lesions. Cancers 13(14):3583. https://doi.org/10.3390/cancers13143583
Shephard AJ, Graham S, Bashir RMS et al (2021) Simultaneous nuclear instance and layer segmentation in oral epithelial dysplasia. In: 2021 IEEE/CVF International Conference on Computer Vision Workshops (ICCVW), pp 552–561. https://doi.org/10.48550/arXiv.2108.13904.
Wang R, Naidu A, Wang Y (2021) Oral cancer discrimination and novel oral epithelial dysplasia stratification using FTIR imaging and machine learning. Diagnostics 11(11):2133. https://doi.org/10.3390/diagnostics11112133
Song B, Sunny S, Uthoff RD et al (2018) Automatic classification of dual-modalilty, smartphone-based oral dysplasia and malignancy images using deep learning. Biomed Opt Express 9(11):5318–5329. https://doi.org/10.1364/boe.9.005318
Aubreville M, Knipfer C, Oetter N et al (2017) Automatic classification of cancerous tissue in laserendomicroscopy images of the oral cavity using deep learning. Sci Rep 7(1). https://doi.org/10.1038/s41598-017-12320-8
Van Staveren H, Van Veen R, Speelman O et al (2000) Classification of clinical autofluorescence spectra of oral leukoplakia using an artificial neural network: a pilot study. Oral Oncol 36(3):286–293. https://doi.org/10.1016/S1368-8375(00)00004-X
Navone R, Burlo P, Pich A et al (2007) The impact of liquid-based oral cytology on the diagnosis of oral squamous dysplasia and carcinoma. Cytopathology 18(6):356–360. https://doi.org/10.1111/j.1365-2303.2006.00402.x
Wang R, Wang Y (2021) Fourier transform infrared spectroscopy in oral cancer diagnosis. Int J Mol Sci 22(3):1206. https://doi.org/10.3390/ijms22031206
Guillaud M, Zhang L, Poh C et al (2008) Potential use of quantitative tissue phenotype to predict malignant risk for oral premalignant lesions. Cancer Res 68(9):3099–3107. https://doi.org/10.1158/0008-5472.Can-07-2113
Issa N, Leonas J, Jham BC et al (2022) Early detection of oral potentially malignant disorders using machine learning: a retrospective pilot study. Gen Dent 70(6):60–64
Birur NP, Song B, Sunny SP et al (2022) Field validation of deep learning based point-of-care device for early detection of oral malignant and potentially malignant disorders. Sci Rep 12(1):14283. https://doi.org/10.1038/s41598-022-18249-x
Farah CS, Janik M, Woo SB et al (2023) Dynamic real-time optical microscopy of oral mucosal lesions using confocal laser endomicroscopy. J Oral Pathol Med 52(6):539–547. https://doi.org/10.1111/jop.13437
Sultan AS, Elgharib MA, Tavares T et al (2020) The use of artificial intelligence, machine learning and deep learning in oncologic histopathology. Journal of Oral Pathology and Medicine. Blackwell Publishing Ltd., pp 849–856. https://doi.org/10.1111/jop.13042
Mahmood H, Shephard A, Hankinson P et al (2023) Development and validation of a multivariable model for prediction of malignant transformation and recurrence of oral epithelial dysplasia. British Journal of Cancer. 129(10):1599–1607. https://doi.org/10.1038/s41416-023-02438-0
Raja Muhammad Saad Bashir, Adam J. Shephard, Hanya Mahmood, et al. (2023) A digital score of peri-epithelial lymphocytic activity predicts malignant transformation in oral epithelial dysplasia. medRxiv: p. 2023.02.14.23285872. https://doi.org/10.1101/2023.02.14.23285872
Azarmehr N, Shephard A, Mahmood H et al (2022) A neural architecture search based framework for segmentation of epithelium, nuclei and oral epithelial dysplasia grading. In: Medical Image Understanding and analysis lecture notes in computer science. Springer, pp 357–370. hhttps://doi.org/10.1007/978-3-031-12053-4_27
Samyukta S, Harini Priya AHR, Kumar SM et al (2022) An evaluation of the prognosticative value of hyalinization in the biological behaviour of oral lesions using image analysis. Asian Pac J Cancer Prev 23(8):2829–2834https://doi.org/10.31557/apjcp.2022.23.8.2829
Adeoye J, Koohi-Moghadam M, Lo AWI et al (2021) Deep learning predicts the malignant-transformation-free survival of oral potentially malignant disorders. Cancers 13(23):6054. https://doi.org/10.3390/cancers13236054
Wu MP, Hsu G, Varvares MA et al (2022) Predicting progression of oral lesions to malignancy using machine learning. Laryngoscope. https://doi.org/10.1002/lary.30285
Mittal P, Condina MR, Klingler-Hoffmann M et al (2021) Cancer tissue classification using supervised machine learning applied to MALDI mass spectrometry imaging. Cancers (Basel). 13(21). https://doi.org/10.3390/cancers13215388
Beuque M, Martin-Lorenzo M, Balluff B et al (2021) Machine learning for grading and prognosis of esophageal dysplasia using mass spectrometry and histological imaging. Comput Biol Med 138:104918. https://doi.org/10.1016/j.compbiomed.2021.104918
Wang R, Naidu A, Wang Y (2021) Oral cancer discrimination and novel oral epithelial dysplasia stratification using FTIR imaging and machine learning. Diagnostics (Basel) 11(11). https://doi.org/10.3390/diagnostics11112133
Rajpurkar P, Lungren MP (2023) The current and future state of AI interpretation of medical images. N Engl J Med 388(21):1981–1990. https://doi.org/10.1056/NEJMra2301725
Alajaji SA, Khoury ZH, Elgharib M et al (2023) Generative adversarial networks in digital histopathology: current applications, limitations, ethical considerations, and future directions. Mod Pathol 37(1):100369. https://doi.org/10.1016/j.modpat.2023.100369
Chen RJ, Lu MY, Chen TY et al (2021) Synthetic data in machine learning for medicine and healthcare. Nat Biomed Eng 5(6):493–497. https://doi.org/10.1038/s41551-021-00751-8
Asan O, Bayrak AE, Choudhury A (2020) Artificial intelligence and human trust in healthcare: focus on clinicians. J Med Internet Res 22(6):e15154. https://doi.org/10.2196/15154
Arrieta AB, Díaz-Rodríguez N, Del Ser J et al (2020) Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf Fusion 58:82–115. https://doi.org/10.1016/j.inffus.2019.12.012
Gunning D, Aha DW (2019) DARPA’s explainable artificial intelligence program. AI Mag 40(2):44–58. https://doi.org/10.1609/aimag.v40i2.2850
Ornes S (2023) Peering inside the black box of AI. Proc Natl Acad Sci 120(22). https://doi.org/10.1073/pnas.2307432120
Samek W, Montavon G, Vedaldi A et al (2019) Explainable AI: interpreting, explaining and visualizing deep learning, Vol. 11700. Springer Nature
Cook KA, Thomas JJ (2005) Illuminating the path: the research and development agenda for visual analytics. Pacific Northwest National Lab. (PNNL), Richland, WA
Nowak S, Rosin M, Stuerzlinger W et al (2021) Visual analytics: a method to explore natural histories of oral epithelial dysplasia. Front Oral Health 2. https://doi.org/10.3389/froh.2021.703874
Quinlan JR (1986) Induction of decision trees. Mach Learn 1(1):81–106. https://doi.org/10.1007/bf00116251
Krishnan MMR, Venkatraghavan V, Acharya UR et al (2012) Automated oral cancer identification using histopathological images: A hybrid feature extraction paradigm. Micron 43(2):352–364. https://doi.org/10.1016/j.micron.2011.09.016
Bilmes JA (1998) A gentle tutorial of the EM algorithm and its application to parameter estimation for Gaussian mixture and hidden Markov models. Int Comput Sci Inst 4(510):126
Altman NS (1992) An introduction to kernel and nearest-neighbor nonparametric regression. Am Stat 46:175–185
Mika S, Ratsch G, Weston J et al (1999) Fisher discriminant analysis with kernels. In: Neural networks for signal processing IX: Proceedings of the 1999 IEEE signal processing society workshop (cat. no. 98th8468). IEEE
Mookiah MRK, P.S., Chandan Chakraborty, Ajoy K Ray, (2011) Brownian motion curve-based textural classification and its application in cancer diagnosis. Anal Quant Cytol Histol 33(3):158–168
Suykens JA, Vandewalle J (1999) Least squares support vector machine classifiers. Neural Process Lett 9:293–300. https://doi.org/10.1023/a:1018628609742
Krishnan MM, Shah P, Choudhary A et al (2011) Textural characterization of histopathological images for oral sub-mucous fibrosis detection. Tissue Cell 43(5):318–330. https://doi.org/10.1016/j.tice.2011.06.005
Krishnan MR, M., M. Pal, S.K. Bomminayuni, et al (2009) Automated classification of cells in sub-epithelial connective tissue of oral sub-mucous fibrosis-an SVM based approach. Comput Biol Med 39(12):1096–1104. https://doi.org/10.1016/j.compbiomed.2009.09.004
Lindner C (2017) Chapter 1 - automated image interpretation using statistical shape models. In: Zheng G, Li S, Székely G (eds) Statistical shape and deformation analysis. Academic Press, pp 3–32. https://doi.org/10.1038/s41598-018-21758-3
Chen L-C, Papandreou G, Kokkinos I et al (2017) Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans Pattern Anal Mach Intell 40(4):834–848. https://doi.org/10.1109/TPAMI.2017.2699184
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III 18. Springer
Guan Q, Wan X, Lu H et al (2019) Deep convolutional neural network Inception-v3 model for differential diagnosing of lymph node in cytological images: a pilot study. Ann Translat Med 7(14):307. https://doi.org/10.21037/atm.2019.06.29
He K, Zhang X, Ren S et al (2016) Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR)
Fati SM, Senan EM, Javed Y (2022) Early diagnosis of oral squamous cell carcinoma based on histopathological images using deep and hybrid learning approaches. Diagnostics (Basel). 12(8). https://doi.org/10.3390/diagnostics12081899
Wang Z, Gao J, Hangyi K et al (2023) ResNet for Histopathologic cancer detection, the deeper, the better? J Data Sci Intell Syst. https://doi.org/10.47852/bonviewJDSIS3202744
Graham S, Quoc DV, Shan EAR et al (2019) HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images. arXiv pre-print server. None arxiv:1812.06499
Neagoe VE, Ciotec AD, Cucu GS (2018) Deep convolutional neural networks versus multilayer perceptron for financial prediction. In: 2018 International Conference on Communications (COMM)
Liu C, Chen L-C, Schroff F et al (2019) Auto-DeepLab: hierarchical neural architecture search for semantic image segmentation. arXiv pre-print server. None arxiv:1901.02985
Huang D-S (1999) Radial basis probabilistic neural networks: model and application. Int J Pattern Recognit Artif Intell 13(7):1083–1101. https://doi.org/10.1142/S0218001499000604
Walczak S, Cerpa N (2003) Artificial neural networks. Encyclopedia of physical science and technology. Academic Press, New York, pp 631–645
Muthu Rama Krishnan M, Shah P, Chakraborty C et al (2012) Statistical analysis of textural features for improved classification of oral histopathological images. J Med Syst 36(2):865–881. https://doi.org/10.1007/s10916-010-9550-8
Alemi Koohbanani N, Jahanifar M, Zamani Tajadin N et al (2020) NuClick: A deep learning framework for interactive segmentation of microscopy images. https://doi.org/10.48550/arXiv.2005.14511.
Acknowledgements
Literature Review Search Strategy: Mary Ann Williams, MSLS. Research & Education Librarian, Health Sciences & Human Services Library, University of Maryland, Baltimore, USA
Funding
This study was not supported by any funding.
Author information
Authors and Affiliations
Contributions
S.A.A. and Z.K. wrote the main manuscript text. S.A.A. prepared the figures and tables. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no conflicts of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
For this type of study, informed consent is not required.
Consent for Publication
For this type of study, consent for publication is not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Alajaji, S.A., Khoury, Z.H., Jessri, M. et al. An Update on the Use of Artificial Intelligence in Digital Pathology for Oral Epithelial Dysplasia Research. Head and Neck Pathol 18, 38 (2024). https://doi.org/10.1007/s12105-024-01643-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12105-024-01643-4