Abstract
The CaMK subfamily of Ser/Thr kinases are regulated by calmodulin interactions with their C-terminal regions. They are exemplified by Ca2+/calmodulin dependent protein kinase 1δ which is known as CaMK1D, CaMKIδ or CKLiK. CaMK1D mediates intracellular signalling downstream of Ca2+ influx and thereby exhibits amplifications of Ca2+signals and polymorphisms that have been implicated in breast cancer and diabetes. Here we report the backbone 1H, 13C, 15N assignments of the 38 kDa human CaMK1D protein in its free state, including both the canonical bi-lobed kinase fold as well as the autoinhibitory and calmodulin binding domains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Biological context
Protein kinases are important mediators of signal transduction, with approximately a 30% of all human proteins being phosphorylated (Cohen 2001). Their deregulation contributes to cancer and many other diseases, and is a major focus of drug discovery efforts (Marsden and Knapp 2008). To date, 518 members have been identified in the human kinome, and are grouped into 10 sub families (Manning et al. 2002). Those that belong to calmodulin dependent kinases (CaMK) group share a similar domain organization. All are activated by binding of Ca2+/calmodulin to their C-terminal regulatory region, releasing the catalytic domains to phosphorylate Ser/Thr residues in protein substrates to alter their functionality (Hook and Means 2001; Soderling and Stull 2001). No resonance assignments for any CaMK member have been reported, limiting analysis of their solution structures and interactions.
The structure of CaMK1D consists of various functional modules. The kinase domain resembles the canonical kinase fold first identified in PKA, which consists of a smaller β-sheet lobe which binds ATP connected by a hinge to the larger α-helical lobe where substrates are recognized. The β-sheet lobe features the P-loop and mobile αC helix, whilst the α-helical lobe presents the catalytic and activation loops (Knighton et al. 1991). Like other CaMK proteins, the kinase domain is negatively regulated by an adjacent C-terminal auto-inhibitory domain (AID), a helix-loop-helix motif which occludes the substrate and ATP binding sites (Goldberg et al. 1996; Swulius and Waxham 2008). The overlapping calmodulin binding domain (CBD) is recognized by calmodulin, which disengages the AID to activate the kinase (Haribabu et al. 1995; Yokokura et al. 1995; Matsushita and Nairn 1998).
Phosphorylation of Thr180 in the activation loop increases CaMK1D activity and is mediated by calcium calmodulin dependent protein kinase kinase (CaMKK). Diverse tissues express CaMK1D including brain, colon, liver, ovary, pancreas, prostate, spleen, testis, and thymus (Ishikawa et al. 2003). Its levels are amplified in basal-like breast tumors, which have a particularly poor clinical outcome. Engineered overexpression of CaMK1D in nontumorgenic cell lines causes increased cell proliferation, migration and invasion, indicating its oncogenic role (Bergamaschi et al. 2008). The subcellular distribution of CaMK1D is predominantly cytoplasmic, but once activated it can translocate to the nucleus (Sakagami et al. 2005). Increased protein expression levels and altered regulation of glucose processing have indicated a role for CaMK1D in diabetes (Fogarty et al. 2014; Haney et al. 2013).
The design of selective inhibitors and drug leads for kinases such as CaMK1D is challenging due to the high degree of conservation within the ATP binding pocket (Vulpetti and Bosotti 2004). NMR spectroscopy provides an efficient route for identifying efficient lead molecules, novel binding pockets and induced conformational changes (Diercks et al. 2001). However, protein kinases are challenging to study by NMR, often yielding poor spectra due to their intrinsic dynamics and large molecular weights. Thus few protein kinases have been assigned (Langer et al. 2004; Vogtherr et al. 2005, 2006; Gelev et al. 2006; Vajpai et al. 2008; Masterson et al. 2009; Xiao et al. 2015; Serimbetov et al. 2017; Sanfelice et al. 2018). Following optimization of solution conditions, the autoinhibited human CaMK1D was found to be amenable to NMR analysis, yielding resolved spectra under physiological buffer conditions. This information has been used to aid in the design of small molecule inhibitors of CaMK1D and development of lead candidates for therapeutic intervention (Fromont et al. 2020).
To enable investigation of CaMK1D’s solution structure and interactions, we have assigned the majority of the backbone resonances of CaMK1D using 2H, 15N and 13C-labelled protein.
Methods and experimental
The construct of CaMK1D comprising the wild type human kinase catalytic domain and autoinhibitory domain (residues 1–333) included an N-terminal His6-tag. The E. coli BL21 DE3 RIPL codon plus strain (Stratagene) was transformed and grown at 37 °C in M9 minimal media supplemented with 30 μg/mL, kanamycin, 34 μg/mL chloramphenicol, 15NH4Cl and 13C6-glucose in 99.9% 2H2O until reaching an O.D 600nm of 0.4. The culture was cooled to 18 °C and induced with 1 mM IPTG for 24 h. The cells were centrifuged at 7000 g for 15 min and resuspended in 50 mM Hepes pH 7.5, 500 mM NaCl, 0.5 mM TCEP, 5 mM imidazole, 0.02% NaN3 supplemented with an EDTA-free complete protease inhibitor cocktail (Roche). The cells were lysed by French press and the resulting cell lysate centrifuged at 75,000×g for 45 min. The supernatant was filtered through a 0.45 µM filter and CaMK1D was purified by Ni2+NTA-affinity chromatography (GE Healthcare). The His6-tag was removed with TEV protease followed again by purification on the Ni2+NTA-affinity column. Final purification was achieved by Superdex-75 (GE Healthcare) size exclusion chromatography in 50 mM Na phosphate pH 7.5, 150 mM NaCl, 0.5 mM TCEP, 0.02% NaN3, which indicated that the protein was monomeric. The CaMK1D sample was exchanged into an NMR buffer containing 50 mM Na phosphate pH 7.0, 75 mM NaCl, 0.5 mM TCEP, 0.02% NaN3 using an Amicon Ultra-15 centrifugal device (Millipore) and concentrated to 1.2 mM in a 600 µL final volume containing 10% 2H2O.
The NMR experiments were performed at 298 K on Varian Inova 800 and 900 MHz NMR spectrometers equipped with triple resonance cryogenic probes with Z-axis pulse field gradients. Backbone assignments were made from TROSY versions of 1H,15 N-HSQC, HNCO, HN(CA)CO, HN(CO)CA, HNCA, HNCACB and HN(COCA)CB (Gardner and Kay 1998). Spectra were processed with NMRPipe (Delaglio et al. 1995) and analysed using CCPN analysis (Vranken et al. 2005).
Extent of assignments and data deposition
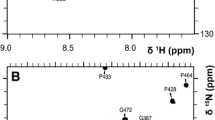
The 1H,15N-TROSY HSQC of the 37.8 kDa deuterated CaMK1D protein in its inactive state is shown (Fig. 1). The backbone HN resonances of 255 residues were assigned out of a possible 322, representing 79% coverage, and including residues from every structural and functional element (Fig. 2). Of those amino acids assigned, 98% include C’, 100% include Cα, and 99% include Cβ chemical shifts. Of all 11 proline residues, 10 were assigned, with 80% including C’, 60% including Cα and 40% including Cβ chemical shifts. The remaining Pro219 could not be assigned as it is N-terminal to another proline. Residues which could not be assigned were 1, 6, 9, 34–35, 56, 63–66, 68–70, 74–77, 82, 84–85, 99–102, 141, 143–144, 146, 164–169, 172, 174, 178–182, 186–187, 199–200, 212–213, 216, 251–254, 260, 303–305, 317–325 and 327. The unassigned residues are mostly found in loop regions, αC helix and C-terminus (Fig. 2). Incomplete assignments in the activation loop and C-terminus also coincided with the disorder and high B-factor values seen for these respective regions in the PDB (2jc6). Resonance assignments in these typically exposed and mobile elements were presumably compromised by peak broadening caused by intermediate or slow time scale dynamics. Assignments were also complicated by peak overlap near the center of the TROSY HSQC spectrum (Fig. 1). Several residues exhibited a second set of weaker or equivalent resonances, including for example Gly103, Gly217 and Asp223 suggesting additional conformers (Fig. 1, green boxes). The Ser residue at the extreme N-terminus which is retained from the TEV site after protease cleavage also could not be assigned. Isolated residue assignments were made for Glu67, Ala83, His142, Leu145 and Tyr 198 based on their unique chemical shifts, intra- and inter-residue correlations in the triple resonance spectra, and the CaMK1D structure.
TROSY HSQC spectrum of deuterated CaMK1D at 1.2 mM concentration in 50 mM Na phosphate pH 7, 75 mM NaCl, 0.5 mM TCEP, 0.02% NaN3 collected at 298 K on a 800 MHz Varian Inova spectrometer. The blue box (denoted A) is an expansion of the crowded central region of the spectrum. The red box (denoted B) further expands the congested central region. Backbone HN peaks are labeled with their assignments. Trp sidechain HN groups are indicated by W-sc. Green boxes highlight examples of residues exhibiting more than one conformation
Backbone assignment coverage of the CaMK1D construct and the secondary structure deduced from the CaMK1D structure (PDBs 2JC6, 6T29, 6T6F). Assigned residues are highlighted in black in the sequence. Dotted green lines represent regions of the construct for which electron density could not be observed in the crystal structure. Solid green lines represent loops, yellow arrows are β-strands and red cylinders are α-helices
The chemical shift values for the 1H, 13C and 15 N resonances of CaMK1D have been deposited at the BioMagResBank (https://www.bmrb.wisc.edu) under accession number.
References
Bergamaschi A, Kim YH, Kwei KA, La Choi Y, Bocanegra M et al (2008) CAMK1D amplification implicated in epithelial-mesenchymal transition in basal-like breast cancer. Mol Oncol 2:327–339
Cohen P (2001) The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur J Biochem 268:5001–5010
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Diercks T, Coles M, Kessler H (2001) Applications of NMR in drug discovery. Curr Opin Chem Biol 5:285–291
Fogarty MP, Cannon ME, Vadlamudi S, Gaulton KJ, Mohlke KL (2014) Identification of a regulatory variant that binds FOXA1 and FOXA2 at the CDC123/CAMK1D type 2 diabetes GWAS locus. PLoS Genet 10:e1004633
Fromont C, Atzori A, Kaur D, Hashmi L, Greco G, Cabanillas A, Nguyen HV, Jones DH, Garzón M, Varela A, Stevenson B, Iacobini GP, Lenoir M, Sundaresan R, Box C, Kumar J, Grant P, Novitskaya V, Morgan J, Sorrell F, Redondo AC, Kramer A, Harris CJ, Leighton B, Vickers SP, Cheetham SC, Kenyon C, Grabowska AM, Overduin M, Berditchevski F, Weston CJ, Knapp S, Fischer PM, Butterworth S (2020) Discovery of highly selective inhibitors of calmodulin-dependent kinases that restore insulin sensitivity in the diet-induced obese in vivo mouse model. J Med Chem (in press)
Gardner KH, Kay LE (1998) The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu Rev Biophys Biomol Struct 27:357–406
Gelev V, Aktas H, Marintchev A, Ito T, Frueh D et al (2006) Mapping of the auto-inhibitory interactions of protein kinase R by nuclear magnetic resonance. J Mol Biol 364:352–363
Goldberg J, Nairn AC, Kuriyan J (1996) Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I. Cell 84:875–887
Haney S, Zhao J, Tiwari S, Eng K, Guey LT, Tien E (2013) RNAi screening in primary human hepatocytes of genes implicated in genome-wide association studies for roles in type 2 diabetes identifies roles for CAMK1D and CDKAL1, among others, in hepatic glucose regulation. PLoS One 8:e64946
Haribabu B, Hook SS, Selbert MA, Goldstein EG, Tomhave ED et al (1995) Human calcium-calmodulin dependent protein kinase I: cDNA cloning, domain structure and activation by phosphorylation at threonine-177 by calcium-calmodulin dependent protein kinase I kinase. EMBO J 14:3679–3686
Hook SS, Means AR (2001) Ca(2+)/CaM-dependent kinases: from activation to function. Annu Rev Pharmacol Toxicol 41:471–505
Ishikawa Y, Tokumitsu H, Inuzuka H, Murata-Hori M, Hosoya H, Kobayashi R (2003) Identification and characterization of novel components of a Ca2+/calmodulin-dependent protein kinase cascade in HeLa cells. FEBS Lett 550:57–63
Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253:407–414
Langer T, Vogtherr M, Elshorst B, Betz M, Schieborr U et al (2004) NMR backbone assignment of a protein kinase catalytic domain by a combination of several approaches: application to the catalytic subunit of cAMP-dependent protein kinase. ChemBioChem 5:1508–1516
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298:1912–1934
Marsden BD, Knapp S (2008) Doing more than just the structure-structural genomics in kinase drug discovery. Curr Opin Chem Biol 12:40–45
Masterson LR, Shi L, Tonelli M, Mascioni A, Mueller MM, Veglia G (2009) Backbone NMR resonance assignment of the catalytic subunit of cAMP-dependent protein kinase A in complex with AMP-PNP. Biomol NMR Assign 3:115–117
Matsushita M, Nairn AC (1998) Characterization of the mechanism of regulation of Ca2+/ calmodulin-dependent protein kinase I by calmodulin and by Ca2+/calmodulin-dependent protein kinase kinase. J Biol Chem 273:21473–21481
Sakagami H, Kamata A, Nishimura H, Kasahara J, Owada Y et al (2005) Prominent expression and activity-dependent nuclear translocation of Ca2+/calmodulin-dependent protein kinase Idelta in hippocampal neurons. Eur J Neurosci 22:2697–2707
Sanfelice D, Koss H, Bunney TD, Thompson GS, Farrell B, Katan M, Breeze AL (2018) NMR backbone assignments of the tyrosine kinase domain of human fibroblast growth factor receptor 3 in apo state and in complex with inhibitor PD173074. Biomol NMR Assign 12:231–235
Serimbetov Z, Baxter NJ, Cliff MJ, Waltho JP (2017) 1H, 15N, 13C backbone resonance assignments of human phosphoglycerate kinase in a transition state analogue complex with ADP, 3-phosphoglycerate and magnesium trifluoride. Biomol NMR Assign 11:251–256
Soderling TR, Stull JT (2001) Structure and regulation of calcium/calmodulin-dependent protein kinases. Chem Rev 101:2341–2352
Swulius MT, Waxham MN (2008) Ca(2+)/calmodulin-dependent protein kinases. Cell Mol Life Sci 65:2637–2657
Vajpai N, Strauss A, Fendrich G, Cowan-Jacob SW, Manley PW et al (2008) Solution conformations and dynamics of ABL kinase-inhibitor complexes determined by NMR substantiate the different binding modes of imatinib/nilotinib and dasatinib. J Biol Chem 283:18292–18302
Vogtherr M, Saxena K, Grimme S, Betz M, Schieborr U et al (2005) NMR backbone assignment of the mitogen-activated protein (MAP) kinase p38. J Biomol NMR 32:175
Vogtherr M, Saxena K, Hoelder S, Grimme S, Betz M et al (2006) NMR characterization of kinase p38 dynamics in free and ligand-bound forms. Angew Chem Int Ed Engl 45:993–997
Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59:687–696
Vulpetti A, Bosotti R (2004) Sequence and structural analysis of kinase ATP pocket residues. Farmaco 59:759–765
Xiao Y, Warner LR, Latham MP, Ahn NG, Pardi A (2015) Structure-based assignment of Ile, Leu, and Val methyl groups in the active and inactive forms of the mitogen-activated protein kinase extracellular signal-regulated kinase 2. Biochemistry 54:4307–4319
Yokokura H, Picciotto MR, Nairn AC, Hidaka H (1995) The regulatory region of calcium/calmodulin-dependent protein kinase I contains closely associated autoinhibitory and calmodulin-binding domains. J Biol Chem 270:23851–23859
Acknowledgements
We thank the staff of the Henry Wellcome Building for Biological NMR Spectroscopy (HWB-NMR), which is funded by the Wellcome Trust, and of NANUC, which is funded by the Canada Foundation for Innovation. This work was funded by the BBSRC and Wellcome Trust [M.O., S.B.] and Campus Alberta Innovation Program chair (M.O.). MT received a BBSRC CASE studentship with GlaxoSmithKline.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tong, M.H.G., Jeeves, M., Rajesh, S. et al. Backbone resonance assignments of the catalytic and regulatory domains of Ca2+/calmodulin-dependent protein kinase 1D. Biomol NMR Assign 14, 221–225 (2020). https://doi.org/10.1007/s12104-020-09950-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-020-09950-x