Abstract
Almost complete sequence specific 1H, 13C and 15N resonance assignments of S114A mutant of UVI31+ from Chlamydomonas reinhardtii are reported. The cDNA of S114A mutant of UVI31+ was cloned from a eukaryotic green algae (C. reinhardtii) and overexpressed in E.coli from where the protein was purified to homogeneity. The point mutation S114A in UVI31+ reduces its DNA endonuclease activity substantially as compared with its wild type. As a prelude to the structural characterization of S114A mutant of UVI31+, we report here complete sequence-specific 1H, 13C and 15N NMR assignments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Biological context
Apoptosis in higher multi-cellular organisms plays an important role in homeostasis, development and defence; however its significance in unicellular organisms remains unclear. Chlamydomonas reinhardtii, a unicellular green alga, has been shown to undergo apoptosis in response to UV-C irradiation (Kim et al. 1997; Moharikar et al. 2006). In order to understand the process of UV mediated apoptosis in C. reinhardtii, we undertook in silico global genome analysis and found a protein, which was identified as UVI31+, one amongst several UV inducible transcripts/proteins and a novel β-lactamase (Moharikar et al. 2006, Rout et al. 2010). UVI31+ protein is also endowed with endonuclease function, in vitro. Cell biological regulation reveals interesting localization changes of UVI31+ protein in Chlamydomonas reinhardtii chloroplast compartment (Shukla et al. 2012).
Chlamydomonas reinhardtii is about 10 μm in diameter that swims with two flagella. They have a cell wall made up of hydroxyproline-rich glycoproteins, a large cup-shaped chloroplast, a large pyrenoid, and an “eyespot” that senses light (Moharikar et al. 2006). It undergoes apoptosis in response to UV-C irradiation (Moharikar et al. 2007). It shows classical hall-marks of animal cell apoptosis and hence can be used as a model system for studying its molecular mechanism in a plant-like environment. Certain candidate molecules were recently identified as to be either UV-regulated or involved in apoptosis. They include apoptosis protease activating factor (APAF), a caspase-3 like protein and a defender against apoptotic death (dad1). All of them exhibited a distinct activation pattern correlating with onset of death following UV irradiation (Moharikar et al. 2006, 2007). One of the putative candidate molecules that is yet to be characterized is the aforementioned UV inducible gene, uvi31+, the focus of the present study.
Uvi31+ was originally isolated from Schizosaccharomyces pombe (Lee et al. 1994), whose expression was unaltered by other DNA damaging or cytotoxic agents. Interestingly, uvi31+ showed no significant sequence homology to the known DNA repair genes. It was observed that uvi31+ transcript increases during normal cell cycle in G1 phase before septation and also during diauxic shift (Kim et al. 1997). A null mutant of uvi31+ from S. pombe showed sensitivity to UV-light, defects in septation and cytokinesis during the resumption of cell division following UV damage-induced cell cycle arrest (Kim et al. 2002).
The uvi31+ protein has structural homology with the BolA protein of Escherichia coli, which was identified by its ability to induce round cell morphology when over-expressed in cells and also following a general stress response (Aldea et al. 1988; Santos et al. 1999; Huynen et al. 2005). Recently, it has been found that uvi31+ exhibited endonuclease activity (Shukla et al. 2012). However, uvi31+ has no structural homology with the known endonucleases. The mutation at S114A site at uvi31+ reduced its endonuclease activity as derived from biochemical analysis (yet to be published). And it has also been speculated that the Ser residue (S114) may be responsible for this activity. With this in the backdrop, we have mutated Ser114 to Ala114 and set out to structurally characterize the mutant of uvi31+ (named as S114Auvi31+) by NMR spectroscopy, to find out whether S114 is involved in endonuclese activity. Towards this goal, we report almost complete sequence specific 1H, 13C and 15N NMR assignments of S114A mutant of UVI31+.
Methods and results
Cloning, over-expression and purification of UVI31+ were carried out as described earlier (Rout et al. 2010). 13C6-glucose or/and 15NH4Cl (Cambridge Isotopes Inc.) were used as the sole source of carbon or/and nitrogen. The purification of the protein was achieved by Ni++-NTA (Ni++- nitrilotriacetate) agarose (Qiagen, Hilgen, Germany). His6-tagged S114A mutant of UVI31+ was eluted with 250 mM imidazole in 50 mM sodium phosphate (pH 7.6), 100 mM NaCl. The eluted fractions were dialyzed overnight against 50 mM sodium phosphate (pH 6.4), 100 mM NaCl.
For NMR experiments, protein samples were prepared in a mixed solvent of 90 % H2O and 10 % 2H2O containing 50 mM sodium phosphate (pH 6.4) and 100 mM NaCl. The protein concentrations were 0.8 mM. NMR experiments were recorded on a Bruker Avance 800 MHz NMR spectrometer equipped with a 5 mm triple-resonance cryogenic probe. Experiments recorded at 298 K with uniformly 15N-labled-S114A mutant of UVI31+ included sensitivity-enhanced 2D [15N–1H]–HSQC using water-flipback for minimizing water saturation (Bax et al. 1991). The 3D experiments recorded with doubly (13C and 15N) labeled samples were, HNCO, HN (CA)CO, CBCANH, and CBCA(CO)NH (Bax et al. 1991; Bax and Grzesiek 1993; Wuthrich et al. 1986), essentially for the assignment of backbone resonances. For the assignment of theThe side chain 1H and 13C resonances, we recordeds were assigned using 3D C(CO)NH and 3D H(CCO)NH (Montelione et al. 1992) and 13C- and 15N-edited NOESY spectra (Muhandiram et al. 1993). NMR data processing was carried out using Bruker Topspin 3.1 software and analyzed with TATAPRO (Atreya et al. 2000, 2002) and CARA (Keller 2002). 1H chemical shifts were referenced with respect to the external standard 2,2-dimethyl-2-silapentene-5-sulfonates (DSS), while 15N and 13C chemical shifts were calibrated indirectly (Edison et al. 1994; Chary and Govil 2008).
Extent of assignment and data deposition
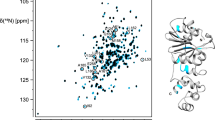
Sequence specific resonance assignments of S114A mutant of UVI31+, which is free of Cys and Trp, could be carried out for nearly all 1H, 13C and 15N spins using a suite of 3D NMR experiments mentioned in Methods. The 1HN and 15N assignments thus derived are shown in the 2D [15N–1H]–HSQC (Fig. 1). A total of 117(1Hi, 13Ci−1,15Ni) correlations are expected from non-proline residues in the 3D-HNCO spectrum of S114A mutant of UVI31+ that has a His-tag of 6 residues. We could however observe 107 distinct correlations and assign almost all of them (97 %) under the experimental conditions described herein. In addition, the side chain 1H and 13C were assigned for most of the residues (88 % of aliphatic 1H resonances and 92 % aliphatic 13C resonances) with an exception of aromatic resonances. The peaks connected by horizontal lines in Fig. 1 belongs to the pairs of side chain NH2 correlations belonging to Asn and Gln residues. The chemical shift data thus obtained has been deposited in the BioMagResBank (http://www.bmrbwisc.edu) under accession number 18567. Sharp [15N–1H] peaks of almost uniform intensity and large dispersion (of 4.4 ppm) of resonances along the 1HN dimension (Fig. 1) revealed that the protein is in a well folded state. The secondary structure elements (Fig. 2) of the protein have been characterized using the well known empirical relations of 13C chemical shifts (∆Cα−∆Cβ) (Spera and Bax 1991; Wishart et al. 1995; Barnwal et al. 2008). The CSI plot (Fig. 2) reveals the presence of both α-helical and β-strand segments separated by short stretches of unstructured elements. Comparison of [15N–1H]–HSQC spectra of (S114A)-UVI31+ with that of wild type-uvi31+, both recorded under similar experimental conditions reveal subtle spectral changes between them, though most of the 15N–1H peaks are unperturbed, suggesting that the overall structural topology of the protein may remain same with substantial conformational changes in the structure particularly near and around the S114A point mutation. Complete 3D structural characterization of the mutant UVI31+ will throw more light on the intricate structural changes caused by the point mutation. This work is in progress.
2D [15N–1H]–HSQC of S114A mutant of UVI31+ at pH 6.4 and 298 K with assignments. The spectrum was recorded with 256 and 1,024 complex points along t1 and t2 dimensions, respectively. The assignments are indicated by the one-letter amino acid code followed by the corresponding sequence number along the protein primary sequence. The peaks connected with horizontal lines are correlations from sidechain NH2 spin pairs belonging to Asn and Gln residues
The secondary structural characterization as derived from the difference between the ∆Cα and ∆Cβ values (∆Cα−∆Cβ) of S114A mutant of UVI31+. The value ∆Cα−∆Cβ for a particular residue i represents the average over the three consecutive residues, i−1, i and i + 1. A stretch, having negative values of ∆Cα−∆Cβ, indicates the presence of extended β-strand whereas positive values indicate a regular α-helical region
References
Aldea M, Hernandez-Chico C, de la Campa AG, Kushner SR, Vicente M (1988) Identification, cloning, and expression of bolA, an ftsZ-dependent morphogene of Escherichia coli. J Bacteriol 170:5169–5176
Atreya HS, Sahu SC, Chary KV, Govil G (2000) A tracked approach for automated NMR assignments in proteins (TATAPRO). J Biomol NMR 17:125–136
Barnwal RP, Rout AK, Atrya HS, Chary KVR (2008) Identification of C-terminal neighbours of amino acid residues without an aliphatic 13Cγ as an aid to NMR assignments in proteins. J Biomol NMR 4:191–197
Bax A, Grzesiek S (1993) Methodological advances in protein NMR. Acc Chem Res 22:131–138
Bax A, Ikura M, Kay LE, Barbato G, Spera S (1991) Multidimensional triple resonance NMR spectroscopy of isotopically uniformly enriched proteins: a powerful new strategy for structure determination. Ciba Found Symp 161:108–119
Chary KVR, Govil G (2008) NMR in biological systems: from molecules to humans. Springer, The Netherlands
Edison AS, Abildgaard F, Westler WM, Mooberry ES, Markley JL (1994) Practical introduction to theory and implementation of multinuclear, multidimensional nuclear magnetic resonance experiments. Methods Enzymol 239:3–79
Huynen MA, Spronk CA, Gabaldon T, Snel B (2005) Combining data from genomes, Y2H and 3D structure indicates that BolA is a reductase interacting with a glutaredoxin. FEBS Lett 579:591–596
Keller R (2002) The computer aided NMR resonance assignment tutorial, CANTINA Verlag Goldau
Kim SH, Kim M, Lee JK, Kim MJ, Jin YH, Seong RH, Hong SH, Joe CO, Park SD (1997) Identification and expression of uvi31+, a UV-inducible gene from Schizosaccharomyces pombe. Environ Mol Mutagen 30:72–81
Kim MJ, Kim HS, Lee JK, Lee CB, Park SD (2002) Regulation of septation and cytokinesis during resumption of cell division requires uvi31+, a UV-inducible gene of fission yeast. Mol Cells 14:425–430
Lee JK, Park EJ, Chung HK, Hong SH, Joe CO, Park SD (1994) Isolation of UV-inducible transcripts from Schizosaccharomyces pombe. Biochem Biophys Res Commun 202:1113–1119
Moharikar S, D’Souza JS, Kulkarni AB, Rao BJ (2006) Apoptotic-like cell death pathway is induced in unicellular chlorophyte Chlamydomonas reinhardtii cells following UV irradiation: detection and functional analyses. J Phycol 42:423–433
Moharikar S, D’Souza JS, Rao BJ (2007) A homologue of the defender against the apoptotic death gene (dad1) in UV-exposed Chlamydomonas cells is downregulated with the onset of programmed cell death. J Biosci 32:261–270
Montelione GT, Lyons BA, Emerson SD, Tashiro M (1992) An efficient triple resonance experiment using carbon-13 isotopic mixing for determining sequence-specific resonance assignment of isotopicallyenriched proteins. J Am Chem Soc 113:5490–5492
Muhandiram DR, Farrow NA, Xu GY, Smallcombe SH, Kay LE (1993) A gradient “C NOESY-HSQC experiment for recording NOESY spectra of “C-labeled proteins dissolved in H2O. J Magn Reson B 102:317–321
Rout AK, Minda R, Peri D, Ramakrishnan V, Bhattacharjee SK, Rao BJ, Chary KVR (2010) Sequence specific 1H, 13C and 15N backbone resonance assignments of UVI31+ from Chlamydomonas reinhardtii. Biomol NMR Assign 4:171–174
Santos JM, Freire P, Vicente M, Arraiano CM (1999) The stationary-phase morphogene bolA from Escherichia coli is induced by stress during early stages of growth. Mol Microbiol 32:789–798
Shukla M, Minda R, Singh H, Tirumani S, Chary KVR and Rao BJ. (2012) UVI31+ is a DNA endonuclease that dynamically localizes to chloroplast pyrenoids in C. reinhardtii. Plos One In press
Spera S, Bax A (1991) Empirical correlation between proteinbackbone conformation and Cα and Cβ 13C nuclear magnetic resonance chemical shifts. J Am Chem Soc 113:5490–5492
Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD (1995) 1H, 13C, and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbour effects. J Biomol NMR 5:67–81
Wuthrich K, Thrich KW, Wuthrich K (1986) NMR of Proteins And Nucleic Acids. Wiley, London
Acknowledgments
The facilities provided by National Facility for High Field NMR at TIFR supported by (DST), Department of Biotechnology (DBT), Council of Scientific and Industrial Research (CSIR) and Tata Institute of Fundamental Research, Mumbai, India are acknowledged. KVRC and BJ thank DST for JC Bose Fellowship. This manuscript is dedicated to Late Prof. Ivano Bertini.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Singh, H., Raghavan, V., Shukla, M. et al. 1H, 13C and 15N resonance assignments of S114A mutant of UVI31+ from Chlamydomonas reinhardtii . Biomol NMR Assign 8, 71–74 (2014). https://doi.org/10.1007/s12104-012-9455-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-012-9455-1