Abstract
Viability and pathogenicity of Gram-negative bacteria is linked to the cytochrome c maturation and the oxidative protein folding systems in the periplasm. The transmembrane reductant conductor DsbD is a unique protein which provides the necessary reducing power to both systems through thiol-disulfide exchange reactions in a complex network of protein–protein interactions. The N-terminal domain of DsbD (nDsbD) is the delivery point of the reducing power originating from cytoplasmic thioredoxin to a variety of periplasmic partners. Here we report 1H, 13C and 15N assignments for resonances of nDsbD in its oxidized and reduced states. These assignments provide the starting point for detailed investigations of the interactions of nDsbD with its protein partners.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Biological Context
In Gram-negative bacteria a large number of extracytoplasmic proteins contain disulfide bonds which are essential for their stability and function (Heras et al. 2009; Riemer et al. 2009). Disulfide bond formation is catalyzed by the Disulfide bond (Dsb) system, comprising five proteins (DsbA-G) located in the periplasm and the inner membrane of the bacterial cell (Porat et al. 2004; Messens and Collet 2006). This system is involved in the formation of disulfide bonds and the isomerization of wrongly formed disulfides. DsbD plays a central role in oxidative folding. It transfers reducing power across the cytoplasmic membrane to DsbC allowing the isomerization of wrongly formed disulfide bonds that leads to correct protein folding (Fabianek et al. 2000; Gleiter and Bardwell 2008). DsbD is also required for the covalent attachment of heme to c-type cytochromes, which are key proteins for the viability of virtually all organisms as they are widely involved in respiratory processes (Thony-Meyer 2002). In c-type cytochromes heme is covalently attached to the CXXCH motif of the apocytochrome via two thioether bonds; the cysteines of this conserved motif need to be in a reduced thiol form (Reid et al. 2001). DsbD is vital for two processes of widespread significance in bacterial life and essential for the survival and virulence of numerous pathogenic organisms. Therefore it is considered a novel target for the development of therapeutics against bacterial infection (Heras et al. 2009).
DsbD consists of three domains, a central transmembrane domain (tmDsbD) flanked by two globular periplasmic domains, the N-terminal (nDsbD) and the C-terminal (cDsbD) domains. Each domain of the protein contains a pair of conserved cysteine residues which are essential for function as the transfer of reductant occurs through a series of thiol:disulfide exchange reactions involving these residues (Katzen and Beckwith 2000). Electrons are transferred from cytoplasmic thioredoxin to tmDsbD, to cDsbD and finally to nDsbD (Gordon et al. 2000; Katzen and Beckwith 2000; Collet et al. 2002; Stirnimann et al. 2006).
nDsbD is the branching point for DsbD-transferred reductant. It acts as a hub which interacts with a variety of protein partners (cDsbD, DsbC, DsbG, CcmG) and thus controls the redox state of the periplasm (Stirnimann et al. 2006). nDsbD is a 16 kDa, globular domain with an immunoglobulin fold, a structural feature not otherwise described for a redox-active protein (Goulding et al. 2002; Haebel et al. 2002). The cysteine pair of this domain (C103 and C109) is located in an unusual CX5C motif and is shielded from solvent exposure by the so-called “cap-loop” residues (residues 66–72) which adopt a more open conformation upon complex formation with the partner proteins (Stirnimann et al. 2006).
X-ray structures for oxidized and reduced nDsbD (Goulding et al. 2002; Haebel et al. 2002; Mavridou et al. 2011) as well as for its covalent mixed disulfide complexes with partner proteins (cDsbD, cDsbD and CcmG) (Haebel et al. 2002; Rozhkova et al. 2004; Stirnimann et al. 2005) have been determined. We have shown in previous studies that NMR can provide important insights into the control of the specificity and reactivity of cDsbD that cannot be obtained from X-ray structures alone (Bushell et al. 2002; Mavridou et al. 2007, 2009, 2011). We now wish to extend these NMR studies to understand how the specificity and reactivity of nDsbD are controlled and to investigate the network of protein–protein interactions of nDsbD, at the residue-specific level. Here we report 1H, 13C and 15N assignments for backbone and sidechain resonances of oxidized and reduced nDsbD.
Methods and experiments
Protein expression and purification
Escherichia coli BL21 (DE3) competent cells were transformed with the expression vector pET22b(+) (Novagen) containing the gene encoding residues L2-V132 of full length E. coli DsbD (N-terminal domain of the protein, nDsbD); the extra residues MDT at the N-terminus and GLVPR at the C-terminus of the protein are a result of the cloning process and the cleavage of a C-terminal His6-tag; assignments for these non-native residues are not reported. Isotopically 15N- and13C/15N-labelled samples were produced using M9 minimal medium enriched with 15NH4Cl and 13C glucose (Cambridge Isotope Laboratories) and supplemented with 1 mM thiamine. Bacteria were grown in volumes of 500 ml to an OD600 of 1.1 in 2 l flasks in the presence of 100 μg.ml−1 carbenicillin from overnight starter cultures (grown at 30°C on Luria–Bertani broth) before addition of isopropyl β-D-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. After overnight incubation the cells were harvested, sphaeroplasted as described (Ausubel et al. 1989), except that EDTA was omitted, and the periplasmic fraction was applied to 10 ml of Fast Flow Chelating Sepharose (Amersham Biosciences) charged with Ni2+. The column was washed with 50 mM Tris–HCl, 150 mM NaCl, 20 mM imidazole, pH 7.5, and the bound protein was eluted with 50 mM Tris–HCl, 150 mM NaCl, 200 mM imidazole, pH 7.5, according to the manufacturer’s instructions. The protein was cleaved using the Sigma Thrombin CleanCleave Kit according to the manufacturer’s instructions.
The purified nDsbD samples contain a mixture of oxidized (with a C103–C109 disulfide) and reduced (without a C103–C109 disulfide) protein. Complete oxidation or reduction of the disulfide bond in nDsbD, prior to NMR experiments, was carried out as follows. 5, 5′-dithiobis-(2-nitrobenzoic acid) (DTNB) was used to oxidize the C103–C109 disulfide bond. 10 mM DTNB was added to the sample and the mixture was incubated at 27°C for 30 min. The C103–C109 disulfide bond in nDsbD samples was reduced using 10 mM dithiothreitol (DTT); complete reduction requires exposure to DTT for at least 12 h (Mavridou et al. 2011).
All constructs were subjected to SDS-PAGE and electrospray ionization mass spectrometry (ES-MS), which was performed using a Micromass Bio-Q II-ZS triple quadrupole mass spectrometer (10 μl protein samples in 1:1 water: acetonitrile, 1% formic acid at a concentration of 20 pmol μl−1 were injected into the electrospray source at a flow rate of 10 μl min−1) to confirm that the proteins were pure and of the expected masses.
NMR spectroscopy
Assignments for oxidized and reduced nDsbD were obtained using uniformly 15N- or 13C/15N-labelled samples of 0.5–1 mM nDsbD in 95% H2O/5% D2O at pH 6.5. All NMR spectra were acquired at 298 K using either a home-built 750 MHz spectrometer which is controlled with GE/Omega software and is equipped with a home-built triple-resonance pulsed-field-gradient probehead or a Bruker Avance 500 MHz spectrometer with a Cryoplatform, equipped with a TCI CryoProbe. Sequential assignments were carried out initially using 15N-labelled nDsbD and 3D 15N-edited TOCSY-HSQC, NOESY-HSQC and HSQC-NOESY-HSQC experiments collected at 750 MHz; analysis of these spectra resulted in nearly complete assignment of the 1HN and 15N resonances for both oxidized and reduced nDsbD. The sequential assignments obtained from the 15N-labelled sample were confirmed with a 3D HNCA experiment collected at 750 MHz and further 13C and 1H assignments were obtained using 3D HNCO, (H)CC(CO)NH and HCCH-TOCSY experiments, collected at 750 MHz, and 3D CBCA(CO)NH, HBHA(CBCACO)NH and HN(CA)CO experiments, collected at 500 MHz.
Extent of assignments and data deposition
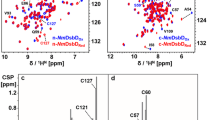
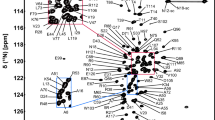
Figure 1a shows the 1H–15N HSQC spectrum of reduced nDsbD. 1HN and 15N backbone resonances for all non-proline residues (L2 to V132 of the native sequence) except E67 have been assigned. In the case of oxidized nDsbD, 1HN and 15N assignments were not obtained for E67, C103, C109 and Y110; it is likely that exchange broadening, due to isomerization of the C103–C109 disulfide bond, is responsible for the absence of peaks corresponding to C103, C109 and Y110. 15N assignments for the 9 proline residues have not been obtained in reduced or oxidized nDsbD. A total of 99.3% of the 1Hα, 99.0% of the 1Hβ, 99.2% of the 13Cα, 98.4% of the 13Cβ and 96.9% of the 13C′ resonances were assigned for reduced nDsbD. For the oxidized state, 97.1% of the 1Hα, 96.2% of the 1Hβ, 97.7% of the 13Cα, 94.4% of the 13Cβ and 94.7% of the 13C′ resonances were assigned. In addition, 60.4% and 45.5% of the remaining sidechain 1H and 13C were assigned for reduced nDsbD, while 52.7% and 42.7% of the remaining sidechain 1H and 13C were assigned for oxidized nDsbD. Most of the missing sidechain assignments correspond to proline and aromatic residues. For reduced nDsbD, 85.9% and 73.0% of the non-proline/non-aromatic sidechain 1H and 13C were assigned. For oxidized nDsbD, 76.7% and 71.1% of the remaining non-proline/non-aromatic sidechain 1H and 13C were assigned. The 1H-15N HSQC spectra of oxidized and reduced nDsbD are overlaid in Fig. 1b and residues showing a combined chemical shift difference (Δδcomb = [Δδ 2HN + 0.1[Δδ15N]2]1/2) of greater than 0.2 ppm are labelled. The combined chemical shift difference is plotted as a function of sequence in Fig. 2. Large chemical shift differences are observed for residues located close to C103 and C109, as would be expected, but also for residues in the cap-loop region which shields the cysteine pair from solvent.
a 750 MHz 1H–15N HSQC spectrum of reduced 15N-labelled nDsbD at pH 6.5, 298 K in 95% H2O/5% D2O. Peak assignments for backbone amides of residues 2-132 of the native sequence are indicated. b Overlay of the 1H–15N HSQC spectra of oxidized (red) and reduced (black) nDsbD. Peaks connected by a solid line indicate the residues for which a large combined chemical shift difference is observed between the two oxidation states. The peaks corresponding to C103 and C109 in reduced nDsbD are indicated with a dashed box; peaks for these residues are not observed in oxidized nDsbD
Combined chemical shift changes (Δδcomb = [Δδ 2HN + 0.1[Δδ15N]2]1/2) observed between the oxidized and reduced states of nDsbD are plotted as a function of amino acid sequence. The red dashed line indicates a combined shift change of greater than 0.2 ppm; these residues are labelled in Fig. 1b
The chemical shift assignments for reduced and oxidized nDsbD have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under the accession numbers 17830 and 17831, respectively.
References
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA (1989) Protein Expression Current Protocols in Molecular Biology. Wiley, New York, pp 16.11–16.18
Bushell KM, Ferguson SF, Redfield C (2002) 1H, 15N and 13C assignments of the carboxy-terminal domain of the transmembrane electron transfer protein DsbD. J Biomol NMR 24:359–360
Collet JF, Riemer J, Bader MW, Bardwell JC (2002) Reconstitution of a disulfide isomerization system. J Biol Chem 277:26886–26892
Fabianek RA, Hennecke H, Thony-Meyer L (2000) Periplasmic protein thiol:disulfide oxidoreductases of Escherichia coli. FEMS Microbiol Rev 24:303–316
Gleiter S, Bardwell JC (2008) Disulfide bond isomerization in prokaryotes. Biochim Biophys Acta 1783:530–534
Gordon EH, Page MD, Willis AC, Ferguson SJ (2000) Escherichia coli DipZ: anatomy of a transmembrane protein disulphide reductase in which three pairs of cysteine residues, one in each of three domains, contribute differentially to function. Mol Microbiol 35:1360–1374
Goulding CW, Sawaya MR, Parseghian A, Lim V, Eisenberg D, Missiakas D (2002) Thiol-disulfide exchange in an immunoglobulin-like fold: structure of the N-terminal domain of DsbD. Biochemistry 41:6920–6927
Haebel PW, Goldstone D, Katzen F, Beckwith J, Metcalf P (2002) The disulfide bond isomerase DsbC is activated by an immunoglobulin-fold thiol oxidoreductase: crystal structure of the DsbC-DsbDalpha complex. EMBO J 21:4774–4784
Heras B, Shouldice SR, Totsika M, Scanlon MJ, Schembri MA, Martin JL (2009) DSB proteins and bacterial pathogenicity. Nat Rev Microbiol 7:215–225
Katzen F, Beckwith J (2000) Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell 103:769–779
Mavridou DAI, Stevens JM, Ferguson SJ, Redfield C (2007) Active-site properties of the oxidized and reduced C-terminal domain of DsbD obtained by NMR spectroscopy. J Mol Biol 370:643–658
Mavridou DAI, Stevens JM, Goddard AD, Willis AC, Ferguson SJ, Redfield C (2009) Control of periplasmic interdomain thiol:disulfide exchange in the transmembrane oxidoreductase DsbD. J Biol Chem 284:3219–3226
Mavridou DAI, Saridakis E, Kritsiligkou P, Goddard AD, Stevens JM, Ferguson SJ, Redfield C (2011) Oxidation state-dependent protein–protein interactions in disulfide cascades. J Biol Chem 286:24943–24956
Messens J, Collet JF (2006) Pathways of disulfide bond formation in Escherichia coli. Int J Biochem Cell Biol 38:1050–1062
Porat A, Cho SH, Beckwith J (2004) The unusual transmembrane electron transporter DsbD and its homologues: a bacterial family of disulfide reductases. Res Microbiol 155:617–622
Reid E, Cole J, Eaves DJ (2001) The Escherichia coli CcmG protein fulfils a specific role in cytochrome c assembly. Biochem J 355:51–58
Riemer J, Bulleid N, Herrmann JM (2009) Disulfide formation in the ER and mitochondria: two solutions to a common process. Science 324:1284–1287
Rozhkova A, Stirnimann CU, Frei P, Grauschopf U, Brunisholz R, Grutter MG, Capitani G, Glockshuber R (2004) Structural basis and kinetics of inter- and intramolecular disulfide exchange in the redox catalyst DsbD. EMBO J 23:1709–1719
Stirnimann CU, Rozhkova A, Grauschopf U, Grutter MG, Glockshuber R, Capitani G (2005) Structural basis and kinetics of DsbD-dependent cytochrome c maturation. Structure 13:985–993
Stirnimann CU, Grutter MG, Glockshuber R, Capitani G (2006) nDsbD: a redox interaction hub in the Escherichia coli periplasm. Cell Mol Life Sci 63:1642–1648
Thony-Meyer L (2002) Cytochrome c maturation: a complex pathway for a simple task? Biochem Soc Trans 30:633–638
Acknowledgments
S.J.F. acknowledge BBSRC funding (grants BBE004865/1 and BBD523019/1). S.J.F., D.A.I.M. and C.R. acknowledge Wellcome Trust funding (grants 079440, 092532 and a Value-in-People Award). D.A.I.M. acknowledges support from the E.P.A. Cephalosporin Fund. L.S.S. acknowledges a BBSRC Vacation Research Bursary.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Mavridou, D.A.I., Stelzl, L.S., Ferguson, S.J. et al. 1H, 13C and 15N resonance assignments for the oxidized and reduced states of the N-terminal domain of DsbD from Escherichia coli . Biomol NMR Assign 6, 163–167 (2012). https://doi.org/10.1007/s12104-011-9347-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-011-9347-9