Abstract
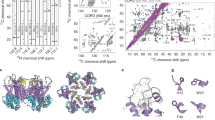
The mature fullerene cone-shaped capsid of the human immunodeficiency virus 1 is composed of about 1,500 copies of the capsid protein (CA). The CA is 231 residues long, and consists of two distinct structural domains, the N-terminal domain and the C-terminal domain (CTD), joined by a flexible linker. The wild type CA exhibits monomer–dimer equilibrium in solution through the CTD–CTD dimerization. This CTD–CTD interaction, together with other intermolecular interdomain interactions, plays significant roles during the assembly of the mature capsid. In addition, CA–CA interactions also play a role in the assembly of the immature virion. The CA also interacts with some host cell proteins within the viral replication cycle. Thus, the capsid protein has been of significant interest as a target for designing inhibitors of assembly of immature virions and mature capsids and inhibitors of its interactions with host cell proteins. However, the equilibrium exhibited by the wild-type CA protein between the monomeric and dimeric states, along with the inherent flexibility from the interdomain linker, have hindered attempts at structural determination by solution NMR and X-ray crystallography methods. In this study, we have utilized a CA protein with W184A and M185A mutations that abolish the dimerization of CA protein as well as its infectivity, but preserve most of the remaining properties of the wild type CA. We have determined the detailed solution structure of the monomeric W184A/M185A-CA protein using 3D-NMR spectroscopy. Here, we present the detailed sequence-specific NMR assignments for this protein.


Similar content being viewed by others
References
Adamson CS, Freed EO (2010) Novel approaches to inhibiting HIV-1 replication. Antiviral Res 85:119–141
Bartels C, Xia T, Billeter M, Guentert P, Wüthrich K (1995) The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR 6:1–10
Byeon I, Meng X, Jung J, Zhao G, Yang R, Ahn J, Shi J, Concel J, Aiken C, Zhang P, Gronenborn A (2009) Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell 139:780–790
Chen B, Tycko R (2010) Structural and dynamical characterization of tubular HIV-1 capsid protein assemblies by solid state nuclear magnetic resonance and electron microscopy. Protein Sci 4:716–730
Delaglio F, Grzesiek S, Vuister GW, Bax A (1995) NMRPipe—a multidimensional spectral processing system based On UNIX pipes. J Biomol NMR 6:277–293
Gamble TR, Yoo S, Vajdos F, von Schwedler U, Worthylake D, Wang H, McCutcheon J, Sundquist W, Hill C (1997) Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849–853
Ganser-Pornillos BK, Cheng A, Yeager M (2007) Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell 131:70–79
Han Y, Ahn J, Concel J, Byeon I, Gronenborn A, Yang J, Polenova T (2010) Solid-state NMR studies of HIV-1 capsid protein assemblies. J Am Chem Soc 6:1976–1987
Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M (2009) X-ray structures of the hexameric building block of the HIV capsid. Cell 137:1282–1292
Tang C, Ndassa Y, Summers MF (2002) Structure of the N-terminal 283-Residue Fragment of the HIV-1 Gag Polyprotein. Nat Struct Biol 9(7):537–543
Wong HC, Shin R, Krishna NR (2008) Solution structure of a double mutant of the carboxy-terminal dimerization domain of the HIV-1 capsid protein. Biochemistry 8:2289–2297
Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ (2007) Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J 26:2218–2226
Acknowledgments
Support of this work by the NIAID Grants 1R21AI081591, 3R21AI081591-02S1, the NCI Grant 1P30 CA13148 that supported the NMR Facility, and the NCRR grant 1S10RR021064-01A1 for the 600 MHz CryoProbe are gratefully acknowledged. Some pilot 3D-NMR studies on a non-His-tag double mutant CA were initially performed at the University of Georgia’s 900 MHz NMR Facility funded by the NIGMS grant GM66340. The authors thank Prof. Peter Prevelige, Jr for the clone of the non-His-tag double mutant CA, and Dr. Clemens Anklin of Bruker Biospin Inc for the SOFAST-HMQC pulse sequence.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Shin, R., Tzou, YM., Wong, H.C. et al. 1H, 15N, and 13C resonance assignments for a monomeric mutant of the HIV-1 capsid protein. Biomol NMR Assign 6, 131–134 (2012). https://doi.org/10.1007/s12104-011-9340-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-011-9340-3