Abstract
The 19.9 kDa C-terminal module (R3) from Azotobacter vinelandii mannronan C5-epimerase AlgE6 has been 13C, 15N isotopically labelled and recombinantly expressed. We report here the 1H, 13C, 15N resonance assignment of AlgE6R3.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Biological context
AlgE6 belongs to a family of seven structurally related alginate epimerases called AlgE1-7 produced by Azotobacter vinelandii (Ertesvåg et al. 1994; Ertesvåg et al. 1995). Alginate is initially produced as poly-β-d-mannuronic acid (M) and alginate epimerases introduce α-l-guluronic acid (G) into the polysaccharide (Hartmann et al. 2002; Campa et al. 2004). Each member of the AlgE-family produces a unique sequence of M and G subunits in the alginate polymer (Ertesvåg and Skjåk-Bræk 1999). All these epimerases consist of two types of structural modules, designated A (~385 amino acids) and R (~150 amino acids). All epimerases contain one N-terminal A-module, and AlgE1 and AlgE3 in addition contain a second such module internally in their sequences (Ertesvåg et al. 1998). The A-modules are the catalytically active parts of the epimerases, but the R-modules strongly enhance this activity although they don’t possess any catalytic activity themselves (Ertesvåg and Valla 1999). The number of R-modules vary from 1 (AlgE4) to 7 (AlgE3), and AlgE6 contains three such modules. Compared to the other two R-modules of AlgE6, R3 has 69 and 64% sequence identity to R1 and R2, respectively. In addition, AlgE6R3 (located C-terminally) contains a predicted C-terminal signal peptide for secretion of the epimerase. The core structures of the R-modules are similar which is also reflected in some conserved chemical shift patterns found in 15N HSQC fingerprint spectra. However, the individual R-modules show quite different affinity for different specifically tailored alginate polymers. Therefore structures of the three R-modules and their affinities to different alginates will allow us to gain a deeper insight into the role of the R-modules in epimerase functionality.
Methods and experiments
The gene coding for the AlgE6R3-module (residues 694–874) was synthesized de novo (GenScript, Piscataway, USA). The sequence was extended by one amino acid (Ala1) for optimal cleavage from the intein tag during purification. The DNA sequence corresponding to AlgE6R3 was cloned into pTYB12 (IMPACT-CN system, New England Biolabs.) using BsmI and XmaI sites, generating pFA13 which codes for a fusion protein consisting of AlgE6R3 and a chitin-binding domain. Uniform labelling of AlgE6R3 (181 amino acids) was achieved by overexpressing the protein in Escherichia coli ER2566 containing the plasmid pFA13. The cells were grown at 37°C to OD600 ~0.8 in M9-medium supplemented with (15NH4)2SO4 (1 g/L), 13C6-d-glucose (2 g/L) (Sigma-Aldrich), 0.2 mM CaCl2 and 200 μg/L ampicillin. Expression was induced by 1 mM ITPG at 15°C and allowed to continue over night. The cells were harvested and resuspended in 20 mM HEPES pH 6.9, 800 mM NaCl, 10 mM CaCl2 and 0.1% Triton X-100 (Sigma-Aldrich). They were then lysed by sonification and the supernatant was loaded on a column with chitin beads (New England BioLabs). The column was washed with 20 mM HEPES pH 6.9, 800 mM NaCl and 5 mM CaCl2. AlgE6R3 was cleaved from the chitin binding tag by incubating the column with the bound fusion protein with 50 mM DTT in 20 mM HEPES pH 6.9, 800 mM NaCl and 5 mM CaCl2 at room temperature for ~16 h, whereafter it could be eluted from the column. The eluted AlgE6R3 was dialysed against 20 mM HEPES, pH 6.9, 25 mM CaCl2.
Samples for NMR studies contained 1.0–1.4 mM AlgE6R3 in 20 mM HEPES buffer, pH 6.9 with 25 mM CaCl2 dissolved in either 90% H2O/10% D2O or 99.9% D2O.
The NMR measurements were performed on a Bruker Avance 600 spectrometer equipped with 5 mm z-gradient TXI (H/C/N) cryogenic probe and on a Bruker DRX 600 spectrometer equipped with 5 mm xyz-gradient TXI (H/C/N) probe. All experiments were performed at 298 K. Proton and carbon chemical shifts were referenced relative to internal 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS); 15N chemical shifts were referenced indirectly to DSS, based on the absolute frequency ratios (Zhang et al. 2003). For the sequence specific backbone- and side-chain assignment, the following experiments were used: 15N-HSQC, 13C-HSQC, HNCO, HN(CA)CO, HNCA, HN(CO)CA, CBCANH, CBCA(CO)NH, HBHANH, HBHA(CO)NH, HCCH-TOCSY and HCCH-COSY. The assignment of aromatic side chains were based on 2D 13C-HSQC and 2D-NOESY, 2D-COSY and 2D-TOCSY recorded on samples dissolved in D2O. The NMR data were recorded and processed with Bruker XWinNMR version 3.5 or Bruker TopSpin 1.3 software and spectral analysis was performed using CARA version 1.4.1/1.8.4 (Keller 2004).
Assignment and data composition
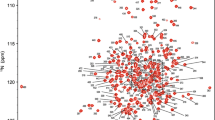
We report here the backbone resonance assignments of the third R-module of AlgE6R3. The 15N-HSQC spectrum of AlgE6R3, together with the assignments of the resonances, is shown in Fig. 1. The backbone and side chain assignment were essentially complete: 96.9% of the backbone HN, Hα, C′, Cα and N atoms, and 95.7% of the side-chain atoms has been assigned. A1 and D2 were not assigned. The amide groups (HN, N) of D14, D47, D118, D119, E142, G143 and A157 could not be found, although other nuclei of these residues were assigned. Except for Hε of R69, none of exchangeable side-chain protons of Arg and Lys residues were assigned. Side-chain amide protons of all Asn and Gln residues were assigned. All aromatic protons were assigned except Hζ of F101. Protonated carbon atoms of aromatic residues were assigned to a large extent.The chemical shift data have been deposited in the BioMagResBank data-base under the accession number 16956.
1H, 15N HSQC spectrum of the 13C, 15N-labelled AlgE6R3 subunit from Azotobacter vinelandii in 90:10 H2O:D2O at pH 6.9, 298 K. Residue numbers are indicated. Side-chain resonances of Asn and Gln residues are connected by lines. Other side-chain amine resonances are indicated with amino acid number and sc
Abbreviations
- DSS:
-
4,4-Dimethyl-4-silapentane-1-sulfonic acid
- DTT:
-
Dithiothreitol
- G:
-
α-l-Guluronic acid
- HEPES:
-
N-2-Hydroxyethylpiperazine-N′-2-ethanesulfonic acid
- IPTG:
-
Isopropyl β-d-1-thiogalactopyranoside
- M:
-
β-d-Mannuronic acid
References
Campa C, Holtan S et al (2004) Biochemical analysis of the processive mechanism for epimerization of alginate by mannuronan C-5 epimerase AlgE4. Biochem J 381(Pt 1):155–164
Ertesvåg H, Skjåk-Bræk G (1999) Modification of alginate using mannuronan C-5-epimerases. In: Bucke C (ed) Carbohydrate biotechnology protocols, vol 10. Humana Press, Totowa, pp 71–78
Ertesvåg H, Valla S (1999) The A modules of the Azotobacter vinelandii mannuronan-C-5-epimerase AlgE1 are sufficient for both epimerization and binding of Ca2+. J Bacteriol 181(10):3033–3038
Ertesvåg H, Doseth B et al (1994) Cloning and expression of an Azotobacter vinelandii mannuronan C-5-epimerase gene. J Bacteriol 176(10):2846–2853
Ertesvåg H, Høidal HK et al (1995) A family of modular type mannuronan C-5-epimerase genes controls alginate structure in Azotobacter vinelandii. Mol Microbiol 16(4):719–731
Ertesvåg H, Høidal HK et al (1998) The Azotobacter vinelandii mannuronan C-5-epimerase AlgE1 consists of two separate catalytic domains. J Biol Chem 273(47):30927–30932
Hartmann M, Holm OB et al (2002) Mode of action of recombinant Azotobacter vinelandii mannuronan C-5 epimerases AlgE2 and AlgE4. Biopolymers 63(2):77–88
Keller RLJ (2004) Optimizing the process of nuclear magnetic resonance spectrum analysis and computer aided resonance assignment. Cantina Verlag, Goldau
Zhang H, Neal S et al (2003) RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR 25(3):173–195
Acknowledgments
This research was supported by the Norwegian Research Council (NFR) via KMB project (182695/I40). EB ad RW acknowledge financial support from the Danish Agency for Science, Technology and Innovation. The NMR laboratory at Aalborg University is supported by the Obel and SparNord foundations.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Buchinger, E., Skjåk-Bræk, G., Valla, S. et al. NMR assignments of 1H, 13C and 15N resonances of the C-terminal subunit from Azotobacter vinelandii mannuronan C5-epimerase 6 (AlgE6R3). Biomol NMR Assign 5, 27–29 (2011). https://doi.org/10.1007/s12104-010-9259-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-010-9259-0