Abstract
The homeobox gene (Hoxa13) codes for a transcription factor protein that binds to AT-rich DNA sequences and controls expression of many important proteins during embryonic morphogenesis. We report complete backbone NMR chemical shift assignments of mouse Hoxa13 DNA binding domain bound to an 11-residue DNA duplex (BMRB no. 16577).
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Biological context
Homeobox (Hox) genes encode a conserved family of transcription factor proteins that are critically important in vertebrate development (Krumlauf 1994). In humans, the Hox genes are distributed into four linkage groups (HOXA, B, C, D) comprising 39 genes located on chromosomes 7, 17, 12, and 2. Recently, mutations in HOXA13 have been associated with Hand-Foot-Genital (HFGS)- and Guttmacher syndromes (GS), autosomal dominant disorders that profoundly affect limb and genitourinary development causing defects in the digits, carpal/tarsal bones, uterus, bladder, Mullerian ducts, and the external genitalia (Innis et al. 2002; Jorgensen et al. 2010; Mortlock and Innis 1997). Analysis of the DNA sequences bound by HOXA13 revealed high affinity for a unique duplex containing a consensus sequence of: 5-AAATAAAA-3′ (Knosp et al. 2007). Characterization of this high affinity sequence in mouse models confirmed that HOXA13 binds to this sequence in vivo and regulates the tissue-specific expression of factors such as bone morphogenetic proteins 2 and 7, to facilitate limb skeletal development (Knosp et al. 2004, 2007).
While it is well-established that mutations affecting the DNA binding domain of HOXA13 cause HFGS and GS, less is known about the amino acids that facilitate HOXA13 DNA binding and/or sequence specificity (Innis et al. 2002; Mortlock and Innis 1997). To gain insight into the molecular basis for the phenotypes present in HFGS and GS, an atomic-resolution structure of HOXA13 bound to duplex DNA is needed to define which amino acids facilitate HOXA13 DNA binding and DNA sequence specificity. Here we report the NMR assignments of the mouse HOXA13 DNA binding domain bound to an 11-residue duplex, forming a complex called, A13DBD-DNA. The NMR assignments of A13DBD-DNA are an important first step toward elucidating the amino acids necessary for sequence specific DNA binding and provide a functional explanation for the loss of HOXA13 function in HFGS and GS.
Methods and experiments
Preparation of HOXA13 DNA binding domain bound to duplex DNA (A13DBD-DNA)
Uniformly 15N-labeled and 13C, 15N-labeled A13DBD were prepared as described previously (Zhang et al. 2009). Two complementary single strands of DNA (1: 5′-CAAATAAAATC-3′ and 2: 5′-GATTTTATTTG-3′ purchased from Bioneer Inc.) were synthesized and HPLC purified. For duplex DNA preparation, the two complementary DNA single strands were dissolved in 1x TE buffer (10 mM TrisHCl, 1 mM EDTA, pH 7.5), mixed in 1:1 M ratio, and annealed at 95°C for 10 min, and then cooled down to room temperature. An aliquot of duplex DNA was added to a stock solution of A13DBD in a 1:1 M ratio and incubated at 15°C for 1 h and the complex was further purified by gel-filtration size-exclusion chromatography (Superdex-75).
NMR spectroscopy
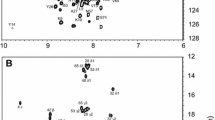
Samples of A13DBD-DNA for NMR analysis were prepared as described above, and exchanged into a buffer containing 20 mM sodium phosphate (pH 6.0) with 5 mM Magnesium chloride and 95% H2O/5% D2O, and finally concentrated to 0.3 ml giving a final protein concentration of 0.5 mM. All NMR experiments were performed at 310 K on a Bruker Avance 800 MHz spectrometer equipped with a four channel interface and triple resonance cryogenic (TCI) probe. The 15N–1H HSQC spectrum (Fig. 1) was recorded with 256 × 2048 complex points for 15N(F1) and 1H(F2). Assignment of backbone resonances was obtained by analyzing the following spectra: HNCA, HNCACB, CBCA(CO)NH, HNCO. The NMR data were processed using NMRPipe and analyzed using Sparky.
Assignments and data deposition
Figure 1 presents the 15N–1H HSQC spectrum of A13DBD-DNA at pH 6.0 to illustrate representative backbone resonance assignments. NMR assignments were based on 3D heteronuclear NMR experiments performed on 13C/15N-labeled A13DBD (residues 1–73). The protein sample in this study consists of 73 native residues without affinity tags or cloning modification. Nearly all non-proline residues exhibited strong backbone amide resonances with uniform intensities in the HSQC spectrum, indicative of a well-defined three-dimensional protein structure. All backbone resonances (1HN, 15N, 13Cα, 13Cβ, and 13CO) were assigned except for the first two residues (G1 and S2) that have very weak NMR intensities. The chemical shift index of each amino acid residue was calculated as described previously (Wishart and Sykes 1994) and revealed three α-helices (α1: T15–T28; α2: K34–N45; α3: E48–K68) and a short β-strand (F31–T33). The protein secondary structure closely resembles the canonical secondary structure and topology seen in other homeobox proteins (Piper et al. 1999). The chemical shift assignments (1H, 15N, 13C) of the A13DBD-DNA complex have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under accession number 16577.
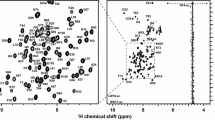
Figure 2 presents the chemical shift perturbation caused by DNA binding to A13DBD. The weighted average chemical shift difference (A13DBD-DNA vs. free protein) was calculated for backbone amide 1H and 15N resonances of each residue. N-terminal residues (K9, R11 and Y14) and residues in the C-terminal helix show the largest chemical shift differences, suggesting that these residues may interact with duplex DNA. Residues I53, Q56, N57 and V60 are all on the same solvent exposed surface of the C-terminal helix and are predicted to make direct sequence-specific contacts inside the major groove analogous to DNA contacts observed in the HoxB1-DNA complex (Piper et al. 1999).
Chemical shift perturbation for A13DBD-DNA. The weighted average chemical shift differences were calculated using \( \Updelta_{av} (HN) = \sqrt {{{\left( {\Updelta H^{2} + \left( {{{\Updelta N} \mathord{\left/ {\vphantom {{\Updelta N} 5}} \right. \kern-\nulldelimiterspace} 5}} \right)^{2} } \right)} \mathord{\left/ {\vphantom {{\left( {\Updelta H^{2} + \left( {{{\Updelta N} \mathord{\left/ {\vphantom {{\Updelta N} 5}} \right. \kern-\nulldelimiterspace} 5}} \right)^{2} } \right)} 2}} \right. \kern-\nulldelimiterspace} 2}} \) where ∆H and ∆N are chemical shift differences between free and DNA-bound states for amide 1H and 15N resonances, respectively. Exposed residues at DNA interface are highlighted in bold. The secondary structure based on CSI is shown at the top (Helix 1: residues 15–28, Helix 2: 34–45, Helix 3: 48–68. A short β-sheet is predicted from 31 to 33)
References
Innis JW, Goodman FR, Bacchelli C, Williams TM, Mortlock DP, Sateesh P, Scambler PJ, McKinnon W, Guttmacher AE (2002) A HOXA13 allele with a missense mutation in the homeobox and a dinucleotide deletion in the promoter underlies Guttmacher syndrome. Hum Mutat 19:573–574
Jorgensen EM, Ruman JI, Doherty L, Taylor HS (2010) A novel mutation of HOXA13 in a family with hand-foot-genital syndrome and the role of polyalanine expansions in the spectrum of Müllerian fusion anomalies. Fertil Steril (in press)
Knosp WM, Scott V, Bachinger HP, Stadler HS (2004) HOXA13 regulates the expression of bone morphogenetic proteins 2 and 7 to control distal limb morphogenesis. Development 131:4581–4592
Knosp WM, Saneyoshi C, Shou S, Bachinger HP, Stadler HS (2007) Elucidation, quantitative refinement, and in vivo utilization of the HOXA13 DNA binding site. J Biol Chem 282:6843–6853
Krumlauf R (1994) Hox genes in vertebrate development. Cell 78:191–201
Mortlock DP, Innis JW (1997) Mutation of HOXA13 in hand-foot-genital syndrome. Nat Genet 15:179–180
Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C (1999) Structure of a HoxB1–Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96:587–597
Wishart DS, Sykes BD (1994) Chemical shifts as a tool for structure determination. Meth Enzymol 239:363–392
Zhang Y, Thornburg CK, Stadler HS, Ames JB (2009) (1)H, (15)N, and (13)C chemical shift assignments of mouse HOXA13 DNA binding domain. Biomol NMR Assign 3:199–201
Acknowledgments
We thank Jeff Walton and Jerry Dallas for technical support and help with NMR experiments. Work supported by NIH grants (EY012347) to J.B.A and (1R01CA131458-01A1) to HSS, and UC Davis NMR facility as well as a Shriners Hospital for Children Research Grant (85800) to HSS.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Zhang, Y., Thornburg, C.K., Stadler, H.S. et al. Backbone chemical shift assignments of mouse HOXA13 DNA binding domain bound to duplex DNA. Biomol NMR Assign 4, 97–99 (2010). https://doi.org/10.1007/s12104-010-9216-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-010-9216-y