Abstract
We have assigned 1H, 15N and 13C resonances of the Bright/ARID DNA-binding domain from the human JARID1C protein, a newly discovered histone demethylase belonging to the JmjC domain-containing protein family.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Biological context
The JARID1C protein (Jumonji/ARID domain-containing protein 1C), also known as SMCX, is a member of the ARID (AT-rich interaction domain) family of DNA-binding proteins, found in fungi, and invertebrate and vertebrate metazoans. The proteins are involved in a variety of biological processes including embryonic development, cell lineage gene regulation and cell cycle control. Although the specific roles of this domain and of ARID-containing proteins in transcriptional regulation are still unclear, they include both positive and negative transcriptional regulation and modification of chromatin structure. JARID1C acts as a histone H3 lysine 4 demethylase involved in transcriptional repression (Iwase et al. 2007). Mutations in JARID1C are a known cause of X-linked mental retardation (XLMR) and epilepsy, suggesting that JARID1C plays a role in human brain function (Tahiliani et al. 2007; Tzschach et al. 2006, Jensen et al. 2005).
The JARID proteins comprise a JmjN domain, a Bright/ARID domain, a JmjC domain, and two PHD-like zinc finger domains. The approximately 100-residue ARID sequence is present in a series of proteins strongly implicated in the regulation of cell growth, development, and tissue-specific gene expression. Although about a dozen ARID-containing proteins can be identified from database searches, to date, only Bright (a regulator of B-cell-specific gene expression), dead ringer (a Drosophila melanogaster gene product required for normal development), and MRF-2 (which represses expression from the cytomegalovirus enhancer) have been analyzed directly in regard to their DNA binding properties (Herrscher et al. 1995; Iwahara et al. 2002; Yuan et al. 1998). Each binds preferentially to AT-rich sites. In contrast, the human SWI–FSNF complex protein p270 shows no sequence preference in its DNA binding activity, thereby demonstrating that AT-rich binding is not an intrinsic property of ARID domains and that ARID family proteins may be involved in a wider range of DNA interactions.
Methods and experiments
Cloning, expression and purification of human JARID1C Bright/ARID domain
The Bright/ARID domain comprising residues 73–188 of the human JARID1C protein (NCBI GenBank GI:11321605, Swissprot, P41229) was cloned into the homemade pNIC28-Bsa4 vector, which adds a TEV-cleavable hexahistidine tag to the N-terminus of the protein, cleavage of which leaves two additional residues, SM on the construct N-terminus. Uniformly 15N- and 15N, 13C-labelled samples of the His-TEV-SM-Bright/ARID domain were grown in E. coli BL21(DE3)-Rosetta cells (Novagen) containing the above plasmid, in M9 minimal medium supplemented with 25 μg/ml kanamycin, 34 μg/ml chloramphenicol and containing 0.5 g/l 15NH4Cl and either 2 g/l (w/v) 12C6-glucose or 13C6-glucose, respectively, as the sole nitrogen and carbon sources. Cultures were grown at 37°C and transferred to 18°C when the OD600 reached 0.6. Expression was induced with 1 mM IPTG and cells were left to grow at 18°C overnight. Cells were harvested by centrifugation, washed with ice-cold 150 mM NaCl and frozen at −80°C. Frozen cell pellets from 2 l culture (∼6.3 g wet mass) were resuspended in a 25 ml lysis buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 5 mM imidazole, 1 mM β-mercaptoethanol, Complete® protease inhibitors EDTA-free (1 tablet/50 ml), 2 mM MgCl2, 10 units Benzonase/ml (>90% purity Novagen), 5 mM arginine and glutamate). The cells were broken by four passes at 16,000 p.s.i. through a high pressure homogeniser (EmulsiFlex-C3/Avestin) followed by centrifugation for 60 min at 60,000g. The supernatant was further clarified by filtration (0.45 μm). Nickel affinity purification was carried out on a workstation Vision (Applied Biosystems). The soluble fraction was loaded on an 8 ml Ni-affinity MC-Poros column with a starting buffer composition of 20 mM Tris–HCl buffer pH 8.0, 500 mM NaCl and 5 mM imidazole at a flow rate of 1 ml/min. Bound protein was eluted with 20 mM Tris–HCl buffer pH 8.0, 500 mM NaCl, 5–500 mM imidazole gradient at 5 ml/min. The extinction at 280 nm was monitored and fractions were collected and analyzed by SDS-PAGE.
The His-tag was cleaved with 1 mg TEV per 40 mg target protein in a dialysis bag (3.5 kDa MW cutoff) and dialysed against 5 l of: 20 mM Tris–HCl 8.0, 500 mM NaCl, 1 mM β-mercaptoethanol, at 15°C overnight. Once cleavage was complete, the sample was supplemented with 5 mM arginine and glutamate and concentrated to 3 ml on a 5 kDa Amicon Ultra-15 centrifuge concentrator. The desired JARID1C Bright/ARID domain was separated from other cleavage products on a 320 ml Superdex 75 column with PBS, pH 7.4, and concentrated using Amicon Ultra-15 concentrators with 5 kDa cutoff. The protein was then exchanged into NMR buffer and concentrated to 1 mM in NMR buffer: 50 mM phosphate, pH 6.0, 50 mM NaCl, 5 mM d-DTT, 0.02% azide, with 10% (v/v) D2O. Protein molecular weights were confirmed by mass spectrometry.
NMR spectroscopy
NMR spectra were acquired at 297 K, using Bruker DRX 600 and DMX 750 spectrometers in standard configuration with triple resonance probes equipped with self-shielded triple axis gradient coils. Spectra for the resonance and NOE assignment were recorded essentially as described in the original references. A 1 mM 13C, 15N-labelled sample of JARID1C in 90% H2O/10% D2O (NMR buffer; pH 6.0) was used for all HN-detected triple resonance experiments; 3D CBCA(CO)NNH, 3D CBCANNH, 3D CC(CO)NNH, 3D H(CCCO)NNH, 3D HBHA(CBCACO)NNH, 3D HNCO, 3D HN(CA)CO, 3D 15N-separated NOESY-HSQC, for 15N T1 and 15N T2 relaxation measurements, for heteronuclear 15N–1H NOE experiments and for a 3D 13C-separated, aliphatic-centred NOESY-HSQC spectrum. The sample was then freeze-dried and redissolved in 100% D2O for acquisition of 3D 13C-separated HMQC-NOESY, 3D HCCH-COSY, and 3D HCCH-TOCSY spectra. Data were processed using the program XWIN-NMR (version 2.6) of Bruker BioSpin GmbH (Rheinstetten, Germany). Assignment of 13C, 15N and 1H resonances was carried out using standard assignment procedures on an Intel Dual Xeon 3GHz PC with the program SPARKY v. 3.1 (Goddard and Kneller).
Extent of assignment and data deposition
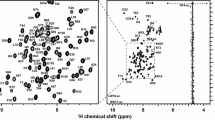
Figure 1 shows the assigned 15N HSQC spectrum of the Bright/ARID domain from the human JARID1C protein. Ninety-four percent of all backbone and sidechain 1H, 15N and 13C resonances were assigned. One hundred percent of Trp Nε1/Hε1, Asn/Gln NH2 and His sidechains were assigned, as well as 75% Phe, 87% Tyr and 30% Arg Nε, Hε sidechains. The assignments were deposited in the BioMagResBank (http://www.bmrb.wisc.edu/) with accession code BMRB-15348.
Abbreviations
- JARID1C:
-
Jumonji/ARID domain-containing protein 1C
- ARID:
-
AT-rich interaction domain
- SMCX:
-
SMCY homologue X-linked
- IPTG:
-
Isopropyl-beta-D-thiogalactopyranoside
References
Goddard TD, Kneller DG, SPARKY 3. University of California, San Francisco. http://www.cgl.ucsf.edu/home/sparky/
Herrscher RF, Kaplan MH, Lelsz DL, Das C, Scheuermann R, Tucker PW (1995) The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Gene Dev 9:3067–3082
Iwahara J, Iwahara M, Daughdrill GW, Ford J, Clubb RT (2002) The structure of the dead ringer-DNA complex reveals how AT-rich interaction domains recognize DNA. EMBO J 21:1197–1209
Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y (2007) The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine demethylases. Cell 128:1077–1088
Jensen LR et al (2005) Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet 76:227–236
Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A Shi Y (2007) The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 447:601–605
Tzschach A et al (2006) Novel JARID1C/SMCX mutations in patients with X-linked mental retardation. Hum Mutat 27:389–394
Yuan YC, Whitson RH, Liu Q, Itakura K, Chen Y (1998) A novel DNA-binding motif shares structural homology to DNA replication and repair nucleases and polymerases. Nat Struct Biol 5:959–964
Acknowledgements
The Structural Genomics Consortium is a registered charity (number 1097737) funded by the Wellcome Trust, GlaxoSmithKline, Genome Canada, the Canadian Institutes of Health Research, the Ontario Innovation Trust, the Ontario Research and Development Challenge Fund and the Canadian Foundation for Innovation. We thank Martina Leidert and Kristina Rehbein for their contribution to sample preparation and Victoria Higman for help with data analysis.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Koehler, C., Bishop, S., Dowler, E.F. et al. Backbone and sidechain 1H, 13C and 15N resonance assignments of the Bright/ARID domain from the human JARID1C (SMCX) protein. Biomol NMR Assign 2, 9–11 (2008). https://doi.org/10.1007/s12104-007-9071-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-007-9071-7