Abstract
Objective
To analyze the infection outbreaks, control measures and outcomes of the outbreak in the NICU of a tertiary care centre in the year 2018.
Methods
This study was conducted in a 30 bedded tertiary care NICU from January 2018 through December 2018. The study design was an Outbreak investigation, based on a program of prospective surveillance for nosocomial infection. All neonates admitted to the NICU formed part of the the study. An Infection Control Quality Improvement (QI) team was available to analyze the infection and initiate response action to outbreaks.
Results
Three outbreaks were reported in the year 2018. The first was in May 2018 and comprised of colonization with rectal Multi-drug resistant gram negative bacilli (MDR GNB). The outbreak was controlled by using Aseptic non-touch technique (ANTT) for fortification of milk and using distilled water for cleaning of diaper area. The second outbreak in August 2018 was Methicillin resistant Staphylococcus aureus (MRSA) whose source was a maternal Lower segment cesarean section (LSCS) wound. The third outbreak in October 2018 was MDR Acinetobacter. The source was from an outborn having the same organism. All infants were in close proximity to the index case. This outbreak was controlled with cohorting, hand hygiene and strengthening of bundle care.
Conclusions
Surveillance aids in early detection and successful control of outbreaks. A systematic search for the source and meticulous containment of spread can successfully control an outbreak.
Similar content being viewed by others
Introduction
Infection outbreaks can have disastrous consequences in the Neonatal intensive care unit (NICU). An outbreak results in the death of an average of 1.5 neonates [1]. It is also found that survivors, after an outbreak, have a longer duration of hospital stay and higher risk of neurodevelopmental sequelae [2]. The cost of treatment also increases with infection. Infection outbreaks contribute to reduction in the morale of healthcare personnel due to maintenance of cohorting and overtime related to care [2].
The definition of an outbreak includes 2 or more isolates of the same species with the same antibiogram within a span of 2 wk. In NICU which screen for colonization, 3 or more infants screened positive for the same Gram negative bacilli (GNB) or a rare GNB that is Extended spectrum beta lactamase (ESBL) should trigger an alert [3].
NICU outbreaks represent one-third of all ICU related outbreaks. The commonest organism reported in recent years include ESBL producing Klebsiella pneumoniae, methicillin resistant Staphylococcus aureus and vancomycin-resistant enterococci. The sources and environmental reservoirs include patients (24.6%), environments (11.7%), personnel (10%), medical equipment devices (9.2%), drugs (5%) and unknown (39.7%) [1].
In India, the Delhi neonatal infection study (DeNIS) reports an alarmingly high prevalence of Acinetobacter. Almost one-third are due to multi-drug resistance (MDR) [4]. It is found that only 26% of NICU outbreaks are investigated [1]. Analysis of the outbreak is important in rapid identification of source and containment of the outbreak. It also aids in strengthening existing preventive measures.
The Outbreak Database provides a large collection of relevant updated information with respect to outbreaks [5].
The objective of this study was to analyze the infection outbreaks, control measures and outcomes of the outbreak in a tertiary care center in the year 2018.
Material and Methods
This study was conducted in a 30 bedded tertiary care NICU from January 2018 through December 2018. The study design was an Outbreak investigation, based on a program of prospective surveillance for nosocomial infection. All neonates admitted to the NICU formed part of the denominator data for the study. There were a total of 40 staff nurses and team of 5 doctors posted in the NICU. An infection control QI team comprising of 3 doctors and 4 nurses were responsible for collection of data and outbreak response investigation. There was a continuous surveillance of infection that had been established as part of a Quality Improvement project in Jan-Sept 2016. The activities of the infection team were:
Collection of data The data included was the total number of babies admitted to NICU, the number of babies on invasive mechanical ventilation, non-invasive ventilation, with central line, intravenous fluid (IV) administration, total parenteral nutrition (TPN) administration, peripheral IV line, IV antibiotics, infants who are nil per oral (NPO).
The data was collected by the NICU staff nurse every morning based on the unit census details. The data was compiled at the end of the month into computer and analyzed once every month. Accuracy of the data was cross-checked on periodic basis and errors were documented along with training of the staff. The details were presented to the nurses and doctors in monthly ward meetings.
A register was maintained for all infants on IV lines and central lines. Hospital acquired Blood stream infection (BSI) and Central line associated blood stream infection (CLABSI) were regarded as sentinel events. They were recorded in a register and a Root cause analysis is done by Hospital Infection Control Committee (HICC) team members and sent to the central HICC. At the end of the month culture data of all infants were collected and analyzed and were presented at the Ward meeting which was attended by all doctors and nurses.
Surveillance of skin and rectal swabs Weekly routine active surveillance for skin (surface culture) and gastrointestinal colonization (stool culture or rectal swab) of Multi-drug resistant organisms (MDROs) [Methicillin resistant Staphylococcus aureus (MRSA) / Vancomycin resistant enterococcus (VRE)/Meropenem resistant GNB] was done for all NICU babies from July 2015. MDRO colonized neonates were isolated and cohorted till their discharge.
MDRO on surface and rectal swabs was defined as any of the following (a) Methicillin resistant Staphylococcus aureus (MRSA) (b) Vancomycin resistant enterococcus (VRE) (c) Gram negative bacillus (GNB) resistant Meropenem.
Infants (3 or more) colonised with the same MDR organism comprised an outbreak. A blood culure with a single MDR was considered an outbreak. The infection control QI team had a meeting once a month to discuss all problems and action that needs to be planned. In case an epidemic was identified, an immediate meeting was called for. All action points that were planned were recorded and executed by the core group members. The action points were communicated to all. Outbreak response measures were recorded. Outbreak-database.com was referenced to aid in identification of source of the epidemic. The outbreak was summarised and presented to all NICU healthcare personnel after it was successfully contained.
The following principles were followed: Rapid identification of cases through surveillance; Cohorting and isolation of cases; Literature search on Outbreak-database.com; Identification of source – Health care personnel (HCP)/ Environment (Depending on organism); Strengthening of hand hygiene, bundle care and environmental process; Communication and education of family members; Determine an antibiotic policy; Follow-up.
Blood cultures were collected prior to antibiotic use in BACTEC bottles. Standard techniques were used. Empirical reports were available by 48 h for necessary action. Environmental samples were collected using standard techniques.
Results
The time periods of the three reported outbreaks and the causative organisms are described in (Table 1). The infants affected in the three outbreaks along with the actions taken and possible source are described in Tables 2, 3 and 4. For the MDR Gram negative bacilli outbreak, 3 or more infants colonised with the same organism was considered an outbreak. During the MRSA outbreak, 3 or infants colonised with MRSA or a single blood culture positive infant with MRSA constituted the outbreak. During the MDR Acinetobacter, the infants were labelled as suspect (with only symptoms), probable [symptoms & C-reactive protein (CRP) positive] and definite (blood culture positive). Out of 3 probable cases, 2 were definite culture positive. Only 1 infant had received colistin without culture positivity.
Discussion
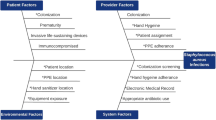
In the NICU, there are multiple risk factors for outbreaks. Prematurity, low birth weight, invasive devices and environmental colonisation are some of the contributing factors. Delayed initiation of enteral nutrition is also a common cause [6, 7].
The outbreak of colonization with MDR Gram negative bacilli was the first of the outbreak described (Table1). A study by Cassetari et al. described breastfeeding was associated with reduced risk for colonisation (OR: 0.22; 95% CI: 0.05–0.99; P = 0.049) [8]. They also isolated the same organism in the hand of a nursing personnel. In authors’ unit, they have a stringent exclusive breastfeeding policy. All efforts are directed to giving every infant mother’s own milk. In the cohort of 11 infants colonized, all were on mother’s own milk. The second factor described by Cassetari et al. is increased use of antibiotics [8]. There was an ongoing antibiotic stewardship QI at that point and all efforts were being made to reduce antibiotic use in the NICU. The outbreak though involved only colonization, required overtime of staff especially to maintain cohorting and isolation. The outbreak was successfully overcome by reinforcing the existing infection control practices and use of aspetic non-touch technique for fortification of milk and use of distilled water to clean the diaper area. However, the cause-effect of the same cannot be proven.
The second outbreak described in August 2018 was the MRSA outbreak (Table 2). The source of the MRSA was identified to be 2 healthcare personnel who had nasal colonization and a mother with MRSA in her LSCS wound swab [She had given Kangaroo mother care (KMC) to her infant]. It is highly debatable whether the healthcare personnel caused the MRSA outbreak or contacted MRSA themselves while taking care of infants during the outbreak. The infants who were colonized were cohorted and given surface cleaning with 0.5% chlorhexidine (bio scrub) for 5 d. The healthcare personnel were allotted to other areas and advised nasal mupirocin for 5 d and chlorhexidine bath. They were screened negative before re-entry to the NICU. Lepelletier et al. found that appropriate cohort isolation of neonates and cohort nursing and improving hand hygiene reduced MRSA transmission. Use of mupirocin in staff and patients was not helpful [9]. Parent to child MRSA transmission has also been reported by other authors [10]. This raises the question of routine parental screening. At this point, this requires further evidence for resource allocation to this intervention. After this outbreak, authors made it a policy to counsel all mothers before KMC and inform any of the HCP if they had a pus discharge of the LSCS wound. All healthcare personnel undergo a screening prior to entry to the NICU.
The third outbreak described, was in October 2018 and was Acinetobacter outbreak (Table 3). The results were devastating and 4 infants expired due to septic shock. The source identified was an extra-mural infant whose blood culture grew Acinetobacter sensitive only to colistin. The key reason for the outbreak was that the blood culture of this infant who expired within 24 h of admission was not followed up. The positive culture was seen only during Root cause analysis of the two deaths of babies no. 2 and 3 (Table 4). Both the deaths occurred before the culture was reported positive. The spread of infection was attributed to compromise in CLABSI bundle care and environmental cleaning practices. This is because the nurse numbers were reduced from a total of 42 to 27. An optimal nurse- infant ratio is necessary to ensure patient safety. All infants were in close proximity to the index case in present cohort. Although mechanical ventilation was a common in all 4 of index infants, only 1 of them was exposed to carbapenem. Acinetobacter is known to thrive in environmental surfaces. Pillay et al. reported source of Acinetobater linked to contaminated suction catheters. De Brito et al. found infected neonates were more exposed to carbapenem and mechanical ventilation [11]. The outbreak response was successful as there were no positive cultures at the institution of the responses. Chan et al. described successful control of Acinetobacter infection with no mortality [12].
Continued surveillance and action plan is needed in order to successfully control outbreaks. Routine screening (skin and rectal swabs) identified the outbreaks early. Measures could be taken to prevent neonatal sepsis and its outcomes. In addition to creating policies, adherence to the policies has to be continuously monitored. However, a culture of ‘zero tolerance” to infection and prompt remedial measures are successful in curtailing outbreaks and saving lives. This is a team effort from the NICU health personnel – nurses, doctors, aides, helpers, parents and also from the HICC and microbiology personnel. The preventive measures to control infection is an ongoing process. The limitation of this study is that authors were unable to do a case-control approach to get a specific risk factor for the outbreak.
Conclusions
Surveillance aids in early detection and successful control of outbreaks. A meticulous search for the source and containment of spread can successfully control an outbreak.
References
Gastmeier P, Loui A, Stamm-Balderjahn S, et al. Outbreaks in neonatal intensive care units - they are not like others. Am J Infect Control. 2007;35:172–6.
Stone PW, Gupta A, Loughrey M, et al. Attributable costs and length of stay of an extended-spectrum beta-lactamase-producing Klebsiella pneumoniae outbreak in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2003;24:601–6.
Anthony M, Bedford-Russell A, Cooper T, et al. Managing and preventing outbreaks of gram-negative infections in UK neonatal units. Arch Dis Child Fetal Neonatal Ed. 2013;98:F549–53.
Investigators of the Delhi Neonatal Infection Study (DeNIS) collaboration. Characterisation and antimicrobial resistance of sepsis pathogens in neonates born in tertiary care centres in Delhi, India: a cohort study. Lancet Glob Health. 2016;4:e752–60.
Vonberg R-P, Weitzel-Kage D, Behnke M, Gastmeier P. Worldwide outbreak database: the largest collection of nosocomial outbreaks. Infection. 2011;39:29–34.
Ramsing BGU, Arpi M, Andersen EA, et al. First outbreak with MRSA in a Danish neonatal intensive care unit: risk factors and control procedures. PLoS One. 2013;8:e66904.
Iosifidis E, Evdoridou I, Agakidou E, et al. Vancomycin-resistant enterococcus outbreak in a neonatal intensive care unit: epidemiology, molecular analysis and risk factors. Am J Infect Control. 2013;41:857–61.
Cassettari VC, da Silveira IR, Dropa M, et al. Risk factors for colonisation of newborn infants during an outbreak of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in an intermediate-risk neonatal unit. J Hosp Infect. 2009;71:340–7.
Lepelletier D, Corvec S, Caillon J, Reynaud A, Rozé J-C, Gras-Leguen C. Eradication of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit: which measures for which success? Am J Infect Control. 2009;37:195–200.
Al-Tawfiq JA. Father-to-infant transmission of community-acquired methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2006;27:636–7.
An outbreak of Acinetobacter baumannii septicemia in a neonatal intensive care unit of a university hospital in Brazil. - PubMed - NCBI [Internet]. Available at: https://www.ncbi.nlm.nih.gov/pubmed/16270122?dopt=Abstract. Accessed 9 Dec 2018.
Chan P-C, Huang L-M, Lin H-C, et al. Control of an outbreak of pandrug-resistant Acinetobacter baumannii colonization and infection in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2007;28:423–9.
Acknowledgements
The authors thank all the faculty, Fellows, Post-graduates and nurses who enabled them to overcome this outbreak.
Author information
Authors and Affiliations
Contributions
BB: Conceptualised the idea and wrote the first draft manuscript; DR: Aided in literature search, edited the first draft manuscript and performed data collection; CLP, SP and SS: Nurses who were involved in data collection and contributed to source identification and control of outbreaks; SR: Reviewed the manuscript and finalised the manuscript. SR will act as guarantor for this paper.
Corresponding author
Ethics declarations
Conflict of Interest
None.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Balachander, B., Rajesh, D., Pinhero, C.L. et al. Response Measures to Infection Outbreaks During the Second Year of Sustenance Phase of Infection Control Quality Improvement. Indian J Pediatr 87, 333–338 (2020). https://doi.org/10.1007/s12098-020-03201-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12098-020-03201-6