Abstract
Purpose
Breast cancer (BrCa) is a predominant type of cancer with a disparate molecular nature. MicroRNAs (miRNAs) have emerged as promising key players in the regulation of pathological processes in BrCa. Proteasome inhibitors (PIs) emerged as promising anticancer agents for several human malignancies, including BrCa, inhibiting the function of the proteasome. Aiming to shed light on the miRNA regulatory effect in BrCa after treatment with PIs, we used two PIs, namely bortezomib and carfilzomib.
Materials and methods
Four BrCa cell lines of distinct molecular subtypes were treated with these PIs. Cell viability and IC50 concentrations were determined. Total RNA was extracted, polyadenylated, and reversely transcribed. Next, the levels of specific miRNAs with a significant role in BrCa were determined using relative quantification, and their regulatory effect was assessed.
Results
High heterogeneity was discovered in the levels of miRNAs in the four cell lines, after treatment. The miRNA levels fluctuate with distinct patterns, in 24, 48, or 72 hours. Interestingly, miR-1-3p, miR-421-3p, and miR-765-3p appear as key molecules, as they were found deregulated, in almost all combinations of cell lines and PIs. In the SK-BR-3 cell line, the majority of the miRNAs were significantly downregulated in treated compared to untreated cells, with miR-21-5p being the only one upregulated. Finally, various significant biological processes, molecular functions, and pathways were predicted to be affected.
Conclusions
The diversity of pathways predicted to be affected by the diversity in miRNA expression after treatment with PIs paves the way for the recognition of new regulatory axes in BrCa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer (BrCa) is the most frequent type of cancer in women, with an expected incidence of over 30% and 15% mortality for 2023 [1]. Novel treatment strategies have greatly improved the survival of patients as the therapeutic approach is personalized and decided based on the molecular characteristics of each patient. The hormone receptors (estrogen and progesterone) status, human epidermal growth factor receptor 2 (HER2) status, and the levels of the proliferation marker Ki-67 are the key components assessed for the stratification of patients into distinct molecular subtypes. Gene mutations in BRCA1, BRCA2, PTEN, TP53, and other genes as well as the expression of specific markers, such as PD-L1, contribute to an improved therapeutic strategy decision [2]. In recent years, numerous drugs have been approved for the treatment of BrCa patients, targeting unique molecules that contribute to the cancerous phenotype, such as Everolimus targeting mTOR and Abemaciclib targeting CDK4/6 [3].
Proteasome inhibitors (PIs) compose a significant class of drugs that regulate protein equilibrium, cell cycle progression, and other biological processes by inhibiting the function of the proteasome [4]. PIs bind to one of the catalytic subunits responsible for the proteasome chymotrypsin-like activity affecting tumor suppressors, cyclin-dependent kinases, metabolic pathways, and other key processes, and may eventually lead to apoptosis [5, 6]. The beneficial effect of PIs is under investigation for the last few years in a series of hematological malignancies and solid tumors revealing promising but, however, puzzling results, because of acquired resistance of cancer cells and side effects in patients. The role of PIs in BrCa has been investigated in numerous research studies and clinical trials focusing mainly on the triple-negative molecular subtype and metastatic BrCa, with a PI being given as a monotherapy or combinational therapy [6, 7]. Despite the fact that PIs are known to affect cell survival and other biological processes by affecting signaling pathways such as the JAKs/STATs and NF-κB pathways, little information exists about their role in BrCa.
MicroRNAs (miRNAs) belong to the class of small non-coding RNAs and have an average length of 22 nucleotides. They appear as fine regulators of gene expression, as well as molecules with a valuable biomarker utility and enormous potential in therapeutic interventions [8, 9]. miRNAs regulate numerous processes that take place in cells including survival, proliferation, differentiation, and metastasis, by sequence similarity with the mRNA target. Their fine tuning depends on their abundance in a specific topology and their involvement in both physiological and pathological states, such as cancer has been well documented [10]. miRNAs appear as key molecules in BrCa [11, 12]. Their role as biomarkers is commonly highlighted [13, 14], and their regulatory potential seems to affect cell viability and death, cell proliferation, tumor aggressiveness, and metastasis [15,16,17]. Their involvement in significant signaling pathways such as the MAPK/ERK [18], PI3K/AKT [19], NF-κB [20], and WNT/β-catenin [21] pathways has also been documented, and their interaction with long non-coding RNAs seems promising as well [22]. Lastly, they appear as molecules with great importance in BrCa therapy, as they have been proposed to contribute to BrCa cell sensitivity to treatment or chemoresistance [23, 24].
Given the significant role of miRNAs in the regulation of BrCa and the previously inadequately studied regulatory potential of PIs at a miRNA post-transcriptional level, we aimed at elucidating the levels of BrCa-significant miRNAs after treatment with PIs, as a first step to pave the way for the recognition and investigation of potential regulatory axes. In this study, specific deregulated miRNAs were identified, after the treatment of BrCa cell lines with PI, and the subsequent effect on biological processes, molecular functions, and pathways was explored. This was elucidated in four BrCa cell lines of distinct molecular subtypes after treatment with two PIs, bortezomib and carfilzomib. To the best of our knowledge, this is the first study that predicts the relative regulatory effect caused by the expression of numerous miRNAs in BrCa, after the treatment with PIs.
Materials and methods
Cell culture conditions
Four well characterized and of distinct molecular subtype BrCa cell lines were purchased from the American Type Culture Collection (ATCC®). The MCF-7 (Luminal A), BT-474 (Luminal B), SK-BR-3 (HER2-positive), and MDA-MB-468 (Triple Negative) cell lines were cultured according to the ATCC guidelines, in a humidified incubator at 37 °C and 5% CO2.
Determination of the IC50 values and documentation of apoptosis after treatment with bortezomib and carfilzomib
Optimal seeding concentrations were determined for the four cell lines, in which the cells would proliferate at a consistent rate achieving a high number of cells in the final 72-hour time point. In the next step, two PIs, bortezomib (Velcade, PS-341) and carfilzomib (Kyprolis, PR-171) were used for the treatment of the four cell lines. Cell viability together with the inhibitor-induced cytotoxicity were assessed in 24, 48, and 72 hours by the Sulforhodamine B assay, using Sulforhodamine B (Invitrogen, Thermo Fisher Scientific), and the Trypan blue exclusion assay, using 0.4% (w/v) Trypan blue solution (Sigma-Aldrich, Merck). The IC50 value, which represents the half-maximal inhibitory concentration of each inhibitor, was determined for each combination of cell line and PI, after performing a wide-range screening of PIs concentrations between 1 and 100 nM. Apoptosis was assessed by the Caspase-3 activity colorimetric assay, following the manufacturer’s instructions (Elabscience, USA). Cell images were obtained with a Carl Zeiss™ Axio Vert.A1 Inverted Microscope (Carl Zeiss, Germany).
Nucleic acid extraction, RNA polyadenylation, and reverse transcription of poly(A) RNA
Total RNA from treated and untreated cells in 24, 48, and 72 hours was extracted using the NucleoZOL (Macherey–Nagel, Germany) reagent and diluted in DEPC-treated H2O. The purity and concentration of total RNA were determined using a BioSpec-nano Micro-volume UV–Vis Spectrophotometer (Shimadju, Kyoto, Japan), and its integrity was assessed electrophoretically in an agarose gel.
Next, 200 ng of total RNA was in vitro polyadenylated using E. coli poly(A) polymerase (New England Biolabs, USA) and first-strand cDNA was synthesized, using an oligo-dT–adapter primer and the M-MLV reverse transcriptase (Invitrogen, Thermo Fisher Scientific). All reactions were performed according to the manufacturer’s instructions in a 96-well Veriti Thermal Cycler (Applied Biosystems, Thermo Fisher Scientific).
Relative quantification of miRNA levels based on real-time qPCR
miRNAs with eminent roles in BrCa were explored in the bibliography and miRNA-specific primers were designed along with primers for the amplification of two reference genes, in order to assess their levels (Table S1). The small nucleolar RNAs SNORD43 and SNORD44 were used as reference genes for the relative quantification of miRNAs using the comparative CT (2−ΔΔCt) method [25]. Real-time qPCR assays followed, using the forward-specific primers, a universal reverse primer, and KAPA™ SYBR® FAST qPCR master mix (2X) (Kapa Biosystems Inc., Woburn, MA, USA), in a total reaction volume of 10 μL. Reactions were performed in a QuantStudio 5 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific Inc.), according to the manufacturer’s protocol, and the prerequisites of the quantitative method were checked. The normalized levels of miRNAs in each sample were calculated using the geometric mean of SNORD43 and SNORD44 levels. The fold change of miRNA levels is presented as a log2 fold change, and a log2 fold change ≥ 1 and ≤ -1 was considered to be significant. Moreover, combinations of BrCa cell lines with PIs treatments, and miRNA expression were clustered using the Heatmapper tool to discover potential similarities between the expression of miRNAs in samples and the miRNA signature of samples [26]. The average linkage clustering method and the Euclidean distance measurement method were chosen for clustering.
Prediction of miRNA targets and affected biological processes, molecular functions, and pathways
The experimentally validated and predicted targets of miRNAs, with significant differences in their expression levels after treatment, were explored using miRTarBase v.9.0, miRDB v.6.0, and TargetScanHuman v.8.0 databases [27,28,29]. Moreover, GO biological processes, GO molecular functions, as well as KEGG pathways in which the experimentally validated and predicted miRNA targets are involved, were identified by custom analysis with Metascape v.3.5 [30].
Results
Optimal seeding concentrations and IC50 values for the treatment of the BrCa cell lines
The optimal seeding concentration in which the cell cultures proliferated at a consistent rate, resulting in a high number of cells at the final 72-hour time point, was determined for each of the four cell lines. These concentrations were 6.00 × 104 cells/mL for the MCF-7 cell line and 1.25 × 105 cells/mL for the BT-474, SK-BR-3, and MDA-MB-468 cell lines.
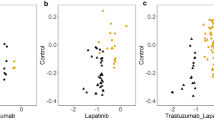
In the next step, a wide-range screening for the approximation of the IC50 values after the treatment of the four cell lines with both bortezomib and carfilzomib was performed (Fig. S1). This screening was performed in 96-well culture plates for 1, 10, and 100 nM of PI in all combinations of cell lines and PIs. Lastly, the IC50 values that indicate the amount of PI that is essential to inhibit the number of viable cells in half in 72 hours were determined, along with the percentage of drug-induced cytotoxicity (Fig. 1). The IC50 values for bortezomib and carfilzomib are 5 nM and 12.5 nM for the MCF-7 cell line, 20 nM and 50 nM for BT-474, 15 nM and 60 nM for SK-BR-3, 10 nM and 40 nM for MDA-MB-468, respectively. Moreover, approximately 50% of cell viability in treated cells can also be observed at 72 hours, in images from the respective cell cultures (Fig. 1). Finally, the presence of PI-induced apoptosis was documented (Fig. S2).
From these data, the MCF-7 cell line which belongs to the Luminal A molecular subtype appears to be more sensitive to the treatment with the two PIs in comparison to the other cell lines. Moreover, in the abovementioned results, treatments with bortezomib appear to be more dose-efficient in all treatment combinations in comparison to treatments with carfilzomib, as bortezomib appears effective in lower concentrations. Furthermore, the drug-induced cytotoxicity appears to be limited in the specified IC50 values.
Expression patterns of miRNAs reflect the high heterogeneity of BrCa molecular subtypes
The relative levels of each miRNA were determined using the comparative CT (2−ΔΔCt) method, with the geometric mean of SNORD43 and SNORD44 serving as a reference. Moreover, the specificity of each amplification product was checked with melt curve analysis. The normalized RQU or fold change values were expressed as log2 fold change for each combination of cell line and inhibitor (Fig. 2 and Fig. 3A).
The expression pattern of miRNAs seems to differ in each combination of cell line and PI which has been used for the relative treatment. Moreover, the expression of miRNAs appears highly heterogeneous over time with specific cases reaching peaks in 24, 48, or 72 hours, respectively, and a restoration of their levels being observed in prolonged time. For example, miR-96-5p levels present a peak in the 24-hour time point whereas miR-16-5p levels present a peak in the 48-hour time point in the BT-474 cells treated with bortezomib. Furthermore, a restoration of the levels of miR-96-5p can be observed in the 72-hour time point in the MCF-7 cells treated with carfilzomib. In many cases, the expression of each miRNA appears to be similar when comparing the two treatment strategies in a specific cell line. However, a limited time difference can be observed in the expression of most miRNAs between the treatments with the two PIs. In addition, the expression of numerous miRNAs, such as let-7a-5p and miR-21-5p in the MDA-MB-468 cell line, appears to not be affected by the treatments in the selected time points.
Interestingly, the majority of miRNAs seem to be considerably downregulated in the treated SK-BR-3 cells in comparison to the relative controls, for both inhibitors. The miR-21-5p appears as an exception in the SK-BR-3 cell line, as it is the only substantially upregulated miRNA after the treatment with bortezomib. Moreover, let-7a-5p, miR-16-5p, and miR-25-3p seem to be significantly downregulated only in the SK-BR-3 cell line. The substantially deregulated miRNAs in each combination of cell line and PI are summarized in Table 1. miR-1-3p, miR-421-3p, and miR-765-3p appear to be significantly deregulated in the majority of treatment combinations, a finding that potentially highlights a considerable role of these miRNAs in the mechanism of action of PIs. Besides these three cases, significantly deregulated miRNAs appear to be different in each combination of cell line and PI.
miRNA expression clustering reveals substantial similarities and differences
In an effort to identify similarities in the expression of miRNAs, clustering was performed using the Heatmapper tool. Clustering of miRNA expression patterns uncovered the similarity in the expression of specific miRNAs in all samples (Fig. 3B). For example, miR-1-3p and miR-421-3p levels appear to derive in a similar time-dependent manner in each cell line studied. Moreover, miR-99a-5p and miR-100-5p, which are members of the same miRNA family and share a similar 5ʹ-seed region, appear to share similar expression patterns as well, among combinations of cell lines and the corresponding treatments.
Furthermore, the clustering of samples according to their miRNA expression patterns revealed the molecular similarity of specific combinations of cell line and PI used for the treatment (Fig. 3C). Interestingly, MCF-7 and BT-474 cell lines that correspond to Luminal A and Luminal B molecular subtypes are characterized by a higher similarity in the miRNA expression profiles. The SK-BR-3 and MDA-MB-468 cell lines that correspond to HER2-positive and triple-negative molecular subtypes appear considerably different regarding their molecular profile, as they are clustered in the two edges of the distribution.
Predicted regulatory effect of miRNA targets resulting from the treatment of BrCa cell lines with bortezomib and carfilzomib
The mRNA targets of the significantly deregulated miRNAs were explored (Table S2), in an effort to investigate the emerging regulatory effect in the BrCa cell lines, after the treatment with the PIs. The top 10 results from each category of affected biological processes, molecular functions, and pathways from the analysis of experimentally validated miRNAs targets, are illustrated for MCF-7 (Fig. 4A), BT-474 (Fig. 4B), SK-BR-3 (Fig. 4C), and MDA-MB-468 (Fig. 4D). Numerous functions, processes, and pathways seem to be altered such as the cell cycle process and cell differentiation which appear to be two of the most deregulated biological processes. Concerning molecular functions, treatment with the two PIs seems to mostly affect numerous molecular binding properties. Moreover, the pathways in cancer seem to be the most enriched pathway in every case and the PI3K/AKT pathway appears to be affected in almost all combinations studied. In addition, great similarities and discordances between cell lines, and the treatment with the two PIs in the same cell line, can be found in this level of analysis, as expected. Lastly, the respective analysis performed in predicted miRNA targets further revealed major discordances between the respective cell line and PI combinations.
Discussion
The significant role of miRNAs in the regulation of BrCa and the remaining ambiguity of miRNA expression regulation by PIs urged us to explore the levels of miRNAs with considerable role in BrCa, after treatment with PIs, in order to recognize potential regulatory axes. Moreover, we chose to include in our study cell lines that belong to the four major molecular subtypes of BrCa in an effort to explore their heterogeneity. The chosen concentrations of the two PIs that were used in this study seem in accordance with previously published data from relevant experiments with a few BrCa cell lines [6, 31]. The MCF-7 cell line appeared to be the most sensitive of the four cell lines that were studied. Moreover, bortezomib appeared more dose-efficient in comparison to carfilzomib, and possessed, in most cases, a higher cytotoxic effect in higher concentrations.
The expression of the studied miRNAs is characterized by great heterogeneity. Moreover, as we study the expression of miRNAs in a broad time range, for transcriptomic purposes, limited or no change in the levels of specific miRNAs is observed. In addition, the restoration of their levels after the effect of the treatment does not allow us to clearly identify an upregulation or downregulation of miRNA levels in the course of time, in most cases. Therefore, a significant deregulation in the levels of each miRNA in treated cells in comparison to the untreated cells is mostly reported. This fact is not a limitation as our goal is to explore the potential regulatory effect after the respective treatments.
The similarity in the expression of specific miRNAs in treatments of a cell line with the two inhibitors may propose a similar mechanism of action, whereas discordances could indicate specific miRNAs as molecules with a controversial role. Interestingly, miR-1-3p, miR-421-3p, and miR-765-3p appear significantly deregulated, and with the same pattern according to clustering, in almost all combinations of treatments. This highlights their significant regulatory impact as they have been found to affect key signaling pathways such as the MAPK, and the NF-κB, and regulate BrCa cell proliferation, tumor growth, metastasis, and chemosensitivity by multiple mechanisms of action [20, 32, 33]. Furthermore, miR-421-3p appears to promote BrCa progression as well as to inhibit BrCa metastasis in another context [17, 34]. Therefore, the study of the regulatory potential of these three miRNAs could unravel novel insight for the mechanism of action of PIs. Three other miRNAs, let-7a-5p, miR-16-5p, and miR-25-3p, appear to be significantly downregulated, and with the same pattern according to clustering, only in the SK-BR-3 cell line. This observation may indicate a fine regulation by these miRNAs in the HER2-positive molecular subtype. However, even though these miRNAs appear as important biomarkers for BrCa [13], and molecules with great regulatory potential, relative scientific literature about their role in HER2-positive BrCa is extremely limited [35]. The other significantly deregulated miRNAs appear to possess a more molecular subtype-specific biomarker utility and regulatory potential, as their levels are considerably deregulated in more specific combinations of BrCa cell line and PI [16, 19, 22, 36,37,38].
Interestingly, the majority of miRNAs seem to be downregulated in the treated SK-BR-3 cells in comparison to the relative controls after treatment with both inhibitors. This appears as an interesting topic requiring further research, as it may reveal a specific mechanism of action of PIs in the HER2-positive molecular subtype. In addition, miR-21-5p appears to be an exception, as it is the only significantly and consistently upregulated miRNA in the SK-BR-3 cell line after treatment with bortezomib. The high levels of this miRNA have been documented to regulate malignant BrCa phenotypes and drug resistance [39, 40]. Therefore, in the context studied, the upregulated miR-21-5p expression after treatment with bortezomib may signify the activation of a bortezomib resistance pathway.
The clustering of samples in relation to the miRNA expression uncovered the molecular similarity of Luminal BrCa molecular subtypes and a high miRNA molecular heterogeneity of the HER2-positive and triple-negative subtypes. This observation validates the importance of miRNAs as biomarkers and molecules with significant regulatory roles with an impact on the molecular heterogeneity of this type of cancer. In addition, according to clustering, miR-99a-5p and miR-100-5p appear to possess similar expression patterns. This observation is important for the significance of the aforementioned results as these two miRNAs belong to the same miRNA family, and therefore, share the same 5ʹ-seed sequence. Therefore, a similar expression in the samples of the study indicates a similar regulatory effect in the studied combinations of BrCa molecular subtypes and treatments.
The regulatory potential of significantly deregulated miRNAs after treatment of BrCa cell lines with PIs uncovered the importance of this type of treatment and its promising therapeutic potential. As revealed from the corresponding analysis, numerous significant pathways, processes, and functions seem to be altered, with the pathways in cancer being the most enriched pathway in every case. Furthermore, the effect of treatments with PIs seems to affect molecular binding properties and to majorly regulate the cell cycle process and cell differentiation. The aforementioned data highlight the significance of treatment with PIs at the miRNA level and subsequently the importance of miRNA regulatory potential in cells.
The specific transcriptional effect of PIs remains largely undiscovered. Most studies focus on multiple myeloma treatment and refer to specific cell properties being affected. Prompted by these properties, specific miRNAs have emerged as regulatory players. For example, miR-145-3p appears to target HDAC4, promoting autophagy and enhancing bortezomib sensitivity in multiple myeloma [41]. Another study indicates miR-497 as a regulator of multiple myeloma cell growth and sensitivity to bortezomib, by the regulation of BCL2 [42]. The effect of PIs on the regulatory potential of miRNAs has not been studied in the context of BrCa, with the only exception being a study that highlights let-7a-5p as an indicator of bortezomib sensitivity [43]. Current bibliography elucidates mainly cell properties and pathways affected after the treatment of BrCa cells, with the PI being provided as a monotherapy or part of combinational therapy, with no reference to miRNAs. Therefore, the innovation of our study is the investigation of the effect of PIs on miRNA expression in BrCa, and the predicted regulatory effect. The investigation of the regulatory effect of miRNAs in different BrCa molecular subtypes is another dynamic innovation with potential applications in the clinic. In addition, relative research for histological types of BrCa could unravel promising results [44]. Furthermore, we chose to investigate the miRNAs levels at numerous time points, in support of the previous bibliography [45], and uncovered the kinetics of the potential regulatory effect of miRNAs. In addition, the heterogeneity in the miRNA expression patterns between the different cell lines that belong to distinct molecular subtypes of BrCa highlights the importance of incorporating the parameter of molecular subtype in relative studies.
The investigation of the regulatory potential of miRNAs with universally deregulated levels in the studied combinations, such as miR-1-3p, as well as of miRNAs with molecular subtype-specific deregulated levels, such as let-7a-5p, could uncover novel mechanisms of action both for miRNAs and PIs in BrCa. Furthermore, the phenomenon of time-dependent differential expression of miRNAs in contrast to constantly downregulated or upregulated levels of miRNAs, such as miR-21-5p after treatment of the SK-BR-3 cell line with bortezomib, appears extremely intriguing. This observation may lead to significant results as a constant deregulation of miRNA levels, up to 72 hours post-treatment, may indicate a molecule as a key player in significant cellular processes and properties, such as the cell cycle and cell sensitivity. Moreover, such a constant expression pattern after treatment indicates specific miRNAs as candidate biomarkers.
In light of the above, and along with the importance of the post-transcriptional miRNA regulation, the validation of regulatory mRNA–miRNA axes after the treatment with PIs is of major importance, to propose interactions with great significance for this type of cancer, and to investigate in depth the therapeutic utility of PIs in BrCa.
Data availability
All data produced in the current study are available from the corresponding author upon reasonable request.
Code availability
The R codes used to visualize the results of functional enrichment analysis are available from the corresponding author upon reasonable request.
References
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics. CA Cancer J Clin. 2023;73(1):17–48. https://doi.org/10.3322/caac.21763.
Loibl S, Poortmans P, Morrow M, Denkert C, Curigliano G. Breast cancer. Lancet. 2021;397(10286):1750–69. https://doi.org/10.1016/S0140-6736(20)32381-3.
Waks AG, Winer EP. Breast cancer treatment: a review. JAMA. 2019;321(3):288–300. https://doi.org/10.1001/jama.2018.19323.
Fricker LD. Proteasome inhibitor drugs. Annu Rev Pharmacol Toxicol. 2020;60:457–76. https://doi.org/10.1146/annurev-pharmtox-010919-023603.
Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr Cancer Drug Targets. 2011;11(3):239–53. https://doi.org/10.2174/156800911794519752.
Shi Y, Yu Y, Wang Z, Wang H, Bieerkehazhi S, Zhao Y, et al. Second-generation proteasome inhibitor carfilzomib enhances doxorubicin-induced cytotoxicity and apoptosis in breast cancer cells. Oncotarget. 2016;7(45):73697–710. https://doi.org/10.18632/oncotarget.12048.
Roeten MSF, Cloos J, Jansen G. Positioning of proteasome inhibitors in therapy of solid malignancies. Cancer Chemother Pharmacol. 2018;81(2):227–43. https://doi.org/10.1007/s00280-017-3489-0.
Katsaraki K, Karousi P, Artemaki PI, Scorilas A, Pappa V, Kontos CK, et al. MicroRNAs: tiny regulators of gene expression with pivotal roles in normal B-cell development and B-cell chronic lymphocytic leukemia. Cancers (Basel). 2021. https://doi.org/10.3390/cancers13040593.
Artemaki PI, Letsos PA, Zoupa IC, Katsaraki K, Karousi P, Papageorgiou SG, et al. The multifaceted role and utility of MicroRNAs in indolent B-cell non-hodgkin lymphomas. Biomedicines. 2021. https://doi.org/10.3390/biomedicines9040333.
Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. https://doi.org/10.1146/annurev-biochem-060308-103103.
Ismail A, El-Mahdy HA, Abulsoud AI, Sallam AM, Eldeib MG, Elsakka EGE, et al. Beneficial and detrimental aspects of miRNAs as chief players in breast cancer: a comprehensive review. Int J Biol Macromol. 2023;224:1541–65. https://doi.org/10.1016/j.ijbiomac.2022.10.241.
Rahman MM, Brane AC, Tollefsbol TO. MicroRNAs and epigenetics strategies to reverse breast cancer. Cells. 2019. https://doi.org/10.3390/cells8101214.
Chen C, Liu X, Chen C, Chen Q, Dong Y, Hou B. Clinical significance of let-7a-5p and miR-21-5p in patients with breast cancer. Ann Clin Lab Sci. 2019;49(3):302–8.
Mavrogiannis AV, Kokkinopoulou I, Kontos CK, Sideris DC. Effect of Vinca alkaloids on the expression levels of microRNAs targeting apoptosis-related genes in breast cancer cell lines. Curr Pharm Biotechnol. 2018;19(13):1076–86. https://doi.org/10.2174/1389201019666181112103204.
Wang Z, Hu S, Li X, Liu Z, Han D, Wang Y, et al. MiR-16-5p suppresses breast cancer proliferation by targeting ANLN. BMC Cancer. 2021;21(1):1188. https://doi.org/10.1186/s12885-021-08914-1.
Shi Y, Zhao Y, Shao N, Ye R, Lin Y, Zhang N, et al. Overexpression of microRNA-96-5p inhibits autophagy and apoptosis and enhances the proliferation, migration and invasiveness of human breast cancer cells. Oncol Lett. 2017;13(6):4402–12. https://doi.org/10.3892/ol.2017.6025.
Hu TB, Chen HS, Cao MQ, Guo FD, Cheng XY, Han ZB, et al. MicroRNA-421 inhibits caspase-10 expression and promotes breast cancer progression. Neoplasma. 2018;65(1):49–54. https://doi.org/10.4149/neo_2018_170306N159.
Zhu X, Qiu J, Zhang T, Yang Y, Guo S, Li T, et al. MicroRNA-188-5p promotes apoptosis and inhibits cell proliferation of breast cancer cells via the MAPK signaling pathway by targeting Rap2c. J Cell Physiol. 2020;235(3):2389–402. https://doi.org/10.1002/jcp.29144.
Zhu B, Chen W, Fu Y, Cui X, Jin L, Chao J, et al. MicroRNA-27a-3p reverses adriamycin resistance by targeting BTG2 and activating PI3K/Akt pathway in breast cancer cells. Onco Targets Ther. 2020;13:6873–84. https://doi.org/10.2147/OTT.S256153.
Chi F, Jin X, Chen L, He G, Han S. TRG16, targeted by miR-765, inhibits breast cancer stem cell-like properties via regulating the NF-kappaB pathway. Mol Cell Biochem. 2022;477(12):2801–16. https://doi.org/10.1007/s11010-022-04480-7.
Wu R, Zhao B, Ren X, Wu S, Liu M, Wang Z, et al. MiR-27a-3p targeting GSK3beta promotes triple-negative breast cancer proliferation and migration through Wnt/beta-catenin pathway. Cancer Manag Res. 2020;12:6241–9. https://doi.org/10.2147/CMAR.S255419.
Lv Y, Lv X, Yang H, Qi X, Wang X, Li C, et al. LncRNA SNHG6/miR-125b-5p/BMPR1B axis: a new therapeutic target for triple-negative breast cancer. Front Oncol. 2021;11: 678474. https://doi.org/10.3389/fonc.2021.678474.
Li F, Miao L, Xue T, Qin H, Mondal S, Thompson PR, et al. Inhibiting PAD2 enhances the anti-tumor effect of docetaxel in tamoxifen-resistant breast cancer cells. J Exp Clin Cancer Res. 2019;38(1):414. https://doi.org/10.1186/s13046-019-1404-8.
Tao S, Bai Z, Liu Y, Gao Y, Zhou J, Zhang Y, et al. Exosomes derived from tumor cells initiate breast cancer cell metastasis and chemoresistance through a MALAT1-dependent mechanism. J Oncol. 2022;2022:5483523. https://doi.org/10.1155/2022/5483523.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. https://doi.org/10.1006/meth.2001.1262.
Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147–53. https://doi.org/10.1093/nar/gkw419.
Huang HY, Lin YC, Cui S, Huang Y, Tang Y, Xu J, et al. miRTarBase update 2022: an informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022;50(D1):D222–30. https://doi.org/10.1093/nar/gkab1079.
Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–31. https://doi.org/10.1093/nar/gkz757.
McGeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM, et al. The biochemical basis of microRNA targeting efficacy. Science. 2019. https://doi.org/10.1126/science.aav1741.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. https://doi.org/10.1038/s41467-019-09234-6.
Wang Y, Rishi AK, Puliyappadamba VT, Sharma S, Yang H, Tarca A, et al. Targeted proteasome inhibition by Velcade induces apoptosis in human mesothelioma and breast cancer cell lines. Cancer Chemother Pharmacol. 2010;66(3):455–66. https://doi.org/10.1007/s00280-009-1181-8.
Yang L, Cai N, Zhao L. MicroRNA-1 regulates the growth and chemosensitivity of breast cancer cells by targeting MEK/ERK pathway. J BUON. 2020;25(5):2215–20.
Jiao Y, Yuan C, Wu H, Li X, Yu J. Oncogenic microRNA-765 promotes the growth and metastasis of breast carcinoma by directly targeting ING4. J Cell Biochem. 2020;121(8–9):3887–900. https://doi.org/10.1002/jcb.29545.
Pan Y, Jiao G, Wang C, Yang J, Yang W. MicroRNA-421 inhibits breast cancer metastasis by targeting metastasis associated 1. Biomed Pharmacother. 2016;83:1398–406. https://doi.org/10.1016/j.biopha.2016.08.058.
Haghi M, Taha MF, Javeri A. Suppressive effect of exogenous miR-16 and miR-34a on tumorigenesis of breast cancer cells. J Cell Biochem. 2019;120(8):13342–53. https://doi.org/10.1002/jcb.28608.
Xia L, Li F, Qiu J, Feng Z, Xu Z, Chen Z, et al. Oncogenic miR-20b-5p contributes to malignant behaviors of breast cancer stem cells by bidirectionally regulating CCND1 and E2F1. BMC Cancer. 2020;20(1):949. https://doi.org/10.1186/s12885-020-07395-y.
Cai Y, Zhang T, Chen G, Liu C. MiR-26a-5p heightens breast cancer cell sensitivity to paclitaxel via targeting flap endonuclease 1. Ann Clin Lab Sci. 2023;53(1):116–25.
Smeets A, Daemen A, Vanden Bempt I, Gevaert O, Claes B, Wildiers H, et al. Prediction of lymph node involvement in breast cancer from primary tumor tissue using gene expression profiling and miRNAs. Breast Cancer Res Treat. 2011;129(3):767–76. https://doi.org/10.1007/s10549-010-1265-5.
Tao L, Wu YQ, Zhang SP. MiR-21-5p enhances the progression and paclitaxel resistance in drug-resistant breast cancer cell lines by targeting PDCD4. Neoplasma. 2019;66(5):746–55. https://doi.org/10.4149/neo_2018_181207N930.
Li J, Huang L, He Z, Chen M, Ding Y, Yao Y, et al. Andrographolide suppresses the growth and metastasis of luminal-like breast cancer by inhibiting the NF-kappaB/miR-21-5p/PDCD4 signaling pathway. Front Cell Dev Biol. 2021;9: 643525. https://doi.org/10.3389/fcell.2021.643525.
Wu H, Liu C, Yang Q, Xin C, Du J, Sun F, et al. MIR145-3p promotes autophagy and enhances bortezomib sensitivity in multiple myeloma by targeting HDAC4. Autophagy. 2020;16(4):683–97. https://doi.org/10.1080/15548627.2019.1635380.
Tian F, Zhan Y, Zhu W, Li J, Tang M, Chen X, et al. MicroRNA-497 inhibits multiple myeloma growth and increases susceptibility to bortezomib by targeting Bcl-2. Int J Mol Med. 2019;43(2):1058–66. https://doi.org/10.3892/ijmm.2018.4019.
Uhr K, der Prager-van Smissen WJC, Heine AAJ, Ozturk B, van Jaarsveld MTM, Boersma AWM, et al. MicroRNAs as possible indicators of drug sensitivity in breast cancer cell lines. PLoS ONE. 2019;14(5):e0216400. https://doi.org/10.1371/journal.pone.0216400.
Papachristopoulou G, Papadopoulos EI, Nonni A, Rassidakis GZ, Scorilas A. Expression analysis of miR-29b in malignant and benign breast tumors: a promising prognostic biomarker for invasive ductal carcinoma with a possible histotype-related expression status. Clin Breast Cancer. 2018;18(4):305–12. https://doi.org/10.1016/j.clbc.2017.11.007.
Bozkurt SB, Ozturk B, Kocak N, Unlu A. Differences of time-dependent microRNA expressions in breast cancer cells. Noncoding RNA Res. 2021;6(1):15–22. https://doi.org/10.1016/j.ncrna.2020.12.001.
Funding
Open access funding provided by HEAL-Link Greece. Part of this work was supported by a research grant from Empirikion Foundation (Athens, Greece). This work was co-financed by the Hellenic Society of Medical Oncology.
Author information
Authors and Affiliations
Contributions
KK performed the experiments, analyzed the data, drafted the manuscript, and produced the figures; CKK had the supervision, designed the study, and critically reviewed the manuscript; GAL and AAT provided the resources and critically reviewed the manuscript; DCS conceived the study and critically reviewed the manuscript; AS conceived the study, provided the resources, and critically reviewed the manuscript; all the authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval and consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Katsaraki, K., Kontos, C.K., Ardavanis-Loukeris, G. et al. Exploring the time-dependent regulatory potential of microRNAs in breast cancer cells treated with proteasome inhibitors. Clin Transl Oncol 26, 1256–1267 (2024). https://doi.org/10.1007/s12094-023-03349-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03349-5