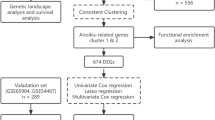
Abstract
Background
Anoikis is a cell death programmed to eliminate dysfunctional or damaged cells induced by detachment from the extracellular matrix. Utilizing an anoikis-based risk stratification is anticipated to understand melanoma's prognostic and immune landscapes comprehensively.
Methods
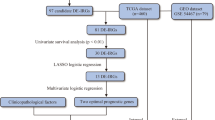
Differential expression genes (DEGs) were analyzed between melanoma and normal skin tissues in The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression data sets. Next, least absolute shrinkage and selection operator, support vector machine–recursive feature elimination algorithm, and univariate and multivariate Cox analyses on the 308 DEGs were performed to build the prognostic signature in the TCGA–melanoma data set. Finally, the signature was validated in GSE65904 and GSE22155 data sets. NOTCH3, PIK3R2, and SOD2 were validated in our clinical samples by immunohistochemistry.
Results
The prognostic model for melanoma patients was developed utilizing ten hub anoikis-related genes. The overall survival (OS) of patients in the high-risk subgroup, which was classified by the optimal cutoff value, was remarkably shorter in the TCGA–melanoma, GSE65904, and GSE22155 data sets. Low-risk patients exhibited low immune cell infiltration and high expression of immunophenoscores and immune checkpoints. They also demonstrated increased sensitivity to various drugs, including dasatinib and dabrafenib. NOTCH3, PIK3R2, and SOD2 were notably associated with OS by univariate Cox analysis in the GSE65904 data set. The clinical melanoma samples showed remarkably higher protein expressions of NOTCH3 (P = 0.003) and PIK3R2 (P = 0.009) than the para-melanoma samples, while the SOD2 protein expression remained unchanged.
Conclusions
In this study, we successfully established a prognostic anoikis-connected signature using machine learning. This model may aid in evaluating patient prognosis, clinical characteristics, and immune treatment modalities for melanoma.









Similar content being viewed by others
Data availability
This study's data generated or examined are comprehensively presented in this article or the supplementary materials. Without reservation, the author is committed to providing the raw data confirming the outcomes of this article to qualified researchers.
Abbreviations
- ML:
-
Machine learning
- GTEx:
-
Genotype-tissue expression
- TCGA:
-
The cancer genome atlas
- SVM–RFE:
-
Support vector machine–recursive feature elimination
- ICPs:
-
Immune checkpoints
- IPSs:
-
Immunophenoscores
- LASSO:
-
Least absolute shrinkage and selection operator
- DEGs:
-
Differentially expressed genes
- DAVID:
-
Database for annotation, visualization and integrated discovery
- TSG101:
-
Tumor susceptibility gene 101
- NQO1:
-
NAD(P)H dehydrogenase (quinone 1)
- EGFR:
-
Epidermal growth factor receptor
- DAPK2:
-
Death-associated protein kinase 2
- CEBPB:
-
CCAAT enhancer binding protein b
- BNIP3:
-
Bcl-2/E1B 19 kDa interacting protein
- ANXA5:
-
Annexin A5
- PIK3R2:
-
Phosphatidylinositol 3-kinase p85beta
- SOD2:
-
Superoxide dismutase 2
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- AUC:
-
Area under the curve
- AJCC:
-
American Joint Committee on Cancer
- ROC:
-
Receiver operating characteristic
- GDSC:
-
Genomics of drug sensitivity in cancer
- IHC:
-
Immunohistochemistry
- HR:
-
Hazard regression
- TME:
-
Tumor microenvironment
- IPSs:
-
Immunophenoscores
- ICPs:
-
Immune checkpoints
- CAF:
-
Cancer-associated fibroblast
- MDSC:
-
Myeloid derived suppressive cell
- M2 TAM:
-
M2 tumor-associated macrophages
- PD-1:
-
Anti-programmed death 1
- CTLA-4:
-
Anti-cytotoxic T lymphocyte-associated antigen 4
- PD-L1:
-
Programmed death ligand 1
- TIM3:
-
Mucin-domain containing-3
- LAG3:
-
Lymphocyte-activation gene-3
References
Dong Z, Yang J, Li L, Tan L, Shi P, Zhang J, et al. FOXO3a-SIRT6 axis suppresses aerobic glycolysis in melanoma. Int J Oncol. 2020;56(3):728–42.
Karimkhani C, Reddy BY, Dellavalle RP, Sundararajan S. Novel therapies for unresectable and metastatic melanoma. BMJ. 2017;359: j5174.
Altonsy MO, Ganguly A, Amrein M, Surmanowicz P, Li SS, Lauzon GJ, et al. Beta3-tubulin is critical for microtubule dynamics, cell cycle regulation, and spontaneous release of microvesicles in human malignant melanoma cells (A375). Int J Mol Sci. 2020;21(5):1656.
Aris M, Barrio MM. Combining immunotherapy with oncogene-targeted therapy: a new road for melanoma treatment. Front Immunol. 2015;6:46.
Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124(4):619–26.
Han HJ, Sung JY, Kim SH, Yun UJ, Kim H, Jang EJ, et al. Fibronectin regulates anoikis resistance via cell aggregate formation. Cancer Lett. 2021;508:59–72.
Adeshakin FO, Adeshakin AO, Afolabi LO, Yan D, Zhang G, Wan X. Mechanisms for modulating anoikis resistance in cancer and the relevance of metabolic reprogramming. Front Oncol. 2021;11: 626577.
Di Micco R, Krizhanovsky V, Baker D, d'Adda di Fagagna F. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat Rev Mol Cell Biol. 2021;22(2): 75–95
Kakavandi E, Shahbahrami R, Goudarzi H, Eslami G, Faghihloo E. Anoikis resistance and oncoviruses. J Cell Biochem. 2018;119(3):2484–91.
Shao Y, Aplin AE. Akt3-mediated resistance to apoptosis in B-RAF-targeted melanoma cells. Cancer Res. 2010;70(16):6670–81.
Madajewski B, Boatman MA, Chakrabarti G, Boothman DA, Bey EA. Depleting tumor-NQO1 potentiates anoikis and inhibits growth of NSCLC. Mol Cancer Res. 2016;14(1):14–25.
Du S, Miao J, Zhu Z, Xu E, Shi L, Ai S, et al. NADPH oxidase 4 regulates anoikis resistance of gastric cancer cells through the generation of reactive oxygen species and the induction of EGFR. Cell Death Dis. 2018;9(10):948.
Du S, Yang Z, Lu X, Yousuf S, Zhao M, Li W, et al. Anoikis resistant gastric cancer cells promote angiogenesis and peritoneal metastasis through C/EBPβ-mediated PDGFB autocrine and paracrine signaling. Oncogene. 2021;40(38):5764–79.
Chen J, Gao F, Liu N. L1CAM promotes epithelial to mesenchymal transition and formation of cancer initiating cells in human endometrial cancer. Exp Ther Med. 2018;15(3):2792–7.
Hsu MY, Yang MH, Schnegg CI, Hwang S, Ryu B, Alani RM. Notch3 signaling-mediated melanoma-endothelial crosstalk regulates melanoma stem-like cell homeostasis and niche morphogenesis. Lab Invest. 2017;97(6):725–36.
Sharma R, Gogoi G, Saikia S, Sharma A, Kalita DJ, Sarma A, et al. BMP4 enhances anoikis resistance and chemoresistance of breast cancer cells through canonical BMP signaling. J Cell Commun Signal. 2022;16(2):191–205.
Adeshakin FO, Adeshakin AO, Liu Z, Cheng J, Zhang P, Yan D, et al. Targeting oxidative phosphorylation-proteasome activity in extracellular detached cells promotes anoikis and inhibits metastasis. Life (Basel). 2021;12(1):42.
Zhou Y, Wang C, Chen Y, Zhang W, Fu Z, Li J, et al. A novel risk model based on anoikis: predicting prognosis and immune infiltration in cutaneous melanoma. Front Pharmacol. 2022;13:1090857.
Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learn. 2002;46(1):389–422.
Ma N, Li J, Lv L, Li C, Li K, Wang B. Bioinformatics evaluation of a novel angiogenesis related genes-based signature for predicting prognosis and therapeutic efficacy in patients with gastric cancer. Am J Transl Res. 2022;14(7):4532–48.
Fan W, Wang D, Li G, Xu J, Ren C, Sun Z, et al. A novel chemokine-based signature for prediction of prognosis and therapeutic response in glioma. CNS Neurosci Ther. 2022;28(12):2090–103.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7): e47.
Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2009.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57.
Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22.
Huang ML, Hung YH, Lee WM, Li RK, Jiang BR. SVM-RFE based feature selection and Taguchi parameters optimization for multiclass SVM classifier. ScientificWorldJournal. 2014;2014: 795624.
Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–45.
Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612.
Maeser D, Gruener RF, Huang RS. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform. 2021;22(6):bbab260.
Guizhen Z, Weiwei Z, Yun W, Guangying C, Yize Z, Zujiang Y. An anoikis-based signature for predicting prognosis in hepatocellular carcinoma with machine learning. Front Pharmacol. 2022;13:1096472.
Sakamoto S, Kyprianou N. Targeting anoikis resistance in prostate cancer metastasis. Mol Aspects Med. 2010;31(2):205–14.
Pierce CJ, Simmons JL, Broit N, Karunarathne D, Ng MF, Boyle GM. BRN2 expression increases anoikis resistance in melanoma. Oncogenesis. 2020;9(7):64.
McCarter MD, Baumgartner J, Escobar GA, Richter D, Lewis K, Robinson W, et al. Immunosuppressive dendritic and regulatory T cells are upregulated in melanoma patients. Ann Surg Oncol. 2007;14(10):2854–60.
Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25(3):486–541.
Simpson CD, Anyiwe K, Schimmer AD. Anoikis resistance and tumor metastasis. Cancer Lett. 2008;272(2):177–85.
González-Llorente L, Santacatterina F, García-Aguilar A, Nuevo-Tapioles C, González-García S, Tirpakova Z, et al. Overexpression of mitochondrial IF1 prevents metastatic disease of colorectal cancer by enhancing anoikis and tumor infiltration of NK cells. Cancers (Basel). 2019;12(1):22.
Wade CA, Kyprianou N. Profiling prostate cancer therapeutic resistance. Int J Mol Sci. 2018;19(3):904.
Chhabra Y, Weeraratna AT. Fibroblasts in cancer: Unity in heterogeneity. Cell. 2023;186(8):1580–609.
Sarkar M, Nguyen T, Gundre E, Ogunlusi O, El-Sobky M, Giri B, et al. Cancer-associated fibroblasts: the chief architect in the tumor microenvironment. Front Cell Dev Biol. 2023;11:1089068.
Mao X, Xu J, Wang W, Liang C, Hua J, Liu J, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer. 2021;20(1):131.
Liu L, Kshirsagar PG, Gautam SK, Gulati M, Wafa EI, Christiansen JC, et al. Nanocarriers for pancreatic cancer imaging, treatments, and immunotherapies. Theranostics. 2022;12(3):1030–60.
Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369(2):122–33.
Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–30.
Lee YH, Martin-Orozco N, Zheng P, Li J, Zhang P, Tan H, et al. Inhibition of the B7–H3 immune checkpoint limits tumor growth by enhancing cytotoxic lymphocyte function. Cell Res. 2017;27(8):1034–45.
Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–62.
Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16(1):64.
Liu Y, Cai P, Wang N, Zhang Q, Chen F, Shi L, et al. Combined blockade of Tim-3 and MEK inhibitor enhances the efficacy against melanoma. Biochem Biophys Res Commun. 2017;484(2):378–84.
Araujo BDLV, Borch A, Hansen M, Draghi A, Spanggaard I, Rohrberg K, et al. Common phenotypic dynamics of tumor-infiltrating lymphocytes across different histologies upon checkpoint inhibition: impact on clinical outcome. Cytotherapy. 2020;22(4):204–13.
Wu J, Liao X, Yu B, Su B. Dasatinib inhibits primary melanoma cell proliferation through morphology-dependent disruption of Src-ERK signaling. Oncol Lett. 2013;5(2):527–32.
Du Four S, Maenhout SK, De Pierre K, Renmans D, Niclou SP, Thielemans K, et al. Axitinib increases the infiltration of immune cells and reduces the suppressive capacity of monocytic MDSCs in an intracranial mouse melanoma model. Oncoimmunology. 2015;4(4): e998107.
Abdel-Wahab O, Klimek VM, Gaskell AA, Viale A, Cheng D, Kim E, et al. Efficacy of intermittent combined RAF and MEK inhibition in a patient with concurrent BRAF- and NRAS-mutant malignancies. Cancer Discov. 2014;4(5):538–45.
Ou C, Liu H, Ding Z, Zhou L. Chloroquine promotes gefitinib-induced apoptosis by inhibiting protective autophagy in cutaneous squamous cell carcinoma. Mol Med Rep. 2019;20(6):4855–66.
Schlegel CR, Georgiou ML, Misterek MB, Stöcker S, Chater ER, Munro CE, et al. DAPK2 regulates oxidative stress in cancer cells by preserving mitochondrial function. Cell Death Dis. 2015;6(3): e1671.
Chua HH, Kameyama T, Mayeda A, and Yeh TH, Cancer-specifically re-spliced TSG101 mRNA promotes invasion and metastasis of nasopharyngeal carcinoma. Int J Mol Sci. 2019;20(3):773.
Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61(18):6669–73.
Peng B, Guo C, Guan H, Liu S, Sun MZ. Annexin A5 as a potential marker in tumors. Clin Chim Acta. 2014;427:42–8.
Siegel D, Franklin WA, Ross D. Immunohistochemical detection of NAD(P)H:quinone oxidoreductase in human lung and lung tumors. Clin Cancer Res. 1998;4(9):2065–70.
Schlager JJ, Powis G. Cytosolic NAD(P)H:(quinone-acceptor)oxidoreductase in human normal and tumor tissue: effects of cigarette smoking and alcohol. Int J Cancer. 1990;45(3):403–9.
Yang Y, Zhang Y, Wu Q, Cui X, Lin Z, Liu S, et al. Clinical implications of high NQO1 expression in breast cancers. J Exp Clin Cancer Res. 2014;33(1):14.
Ma Y, Kong J, Yan G, Ren X, Jin D, Jin T, et al. NQO1 overexpression is associated with poor prognosis in squamous cell carcinoma of the uterine cervix. BMC Cancer. 2014;14:414.
Li Z, Zhang Y, Jin T, Men J, Lin Z, Qi P, et al. NQO1 protein expression predicts poor prognosis of non-small cell lung cancers. BMC Cancer. 2015;15:207.
Park SJ, Zhao H, Spitz MR, Grossman HB, Wu X. An association between NQO1 genetic polymorphism and risk of bladder cancer. Mutat Res. 2003;536(1–2):131–7.
Thapa D, Huang SB, Muñoz AR, Yang X, Bedolla RG, Hung CN, et al. Attenuation of NAD[P]H:quinone oxidoreductase 1 aggravates prostate cancer and tumor cell plasticity through enhanced TGFβ signaling. Commun Biol. 2020;3:12.
Du Q, Tan Z, Shi F, Tang M, Xie L, Zhao L, et al. PGC1α/CEBPB/CPT1A axis promotes radiation resistance of nasopharyngeal carcinoma through activating fatty acid oxidation. Cancer Sci. 2019;110(6):2050–62.
Jerhammar F, Ceder R, Garvin S, Grénman R, Grafström RC, Roberg K. Fibronectin 1 is a potential biomarker for radioresistance in head and neck squamous cell carcinoma. Cancer Biol Ther. 2010;10(12):1244–51.
Guerzoni C, Bardini M, Mariani SA, Ferrari-Amorotti G, Neviani P, Panno ML, et al. Inducible activation of CEBPB, a gene negatively regulated by BCR/ABL, inhibits proliferation and promotes differentiation of BCR/ABL-expressing cells. Blood. 2006;107(10):4080–9.
Termini L, Fregnani JH, Boccardo E, da Costa WH, Longatto-Filho A, Andreoli MA, et al. SOD2 immunoexpression predicts lymph node metastasis in penile cancer. BMC Clin Pathol. 2015;15:3.
Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13(3):184–99.
Vartanian A, Gatsina G, Grigorieva I, Solomko E, Dombrovsky V, Baryshnikov A, et al. The involvement of Notch signaling in melanoma vasculogenic mimicry. Clin Exp Med. 2013;13(3):201–9.
Ivan C, Hu W, Bottsford-Miller J, Zand B, Dalton HJ, Liu T, et al. Epigenetic analysis of the Notch superfamily in high-grade serous ovarian cancer. Gynecol Oncol. 2013;128(3):506–11.
Lai IC, Shih PH, Yao CJ, Yeh CT, Wang-Peng J, Lui TN, et al. Elimination of cancer stem-like cells and potentiation of temozolomide sensitivity by Honokiol in glioblastoma multiforme cells. PLoS ONE. 2015;10(3): e0114830.
Wang J, Cai S, Xiong Q, Weng D, Wang Q, Ma Z. PIK3R2 predicts poor outcomes for patients with melanoma and contributes to the malignant progression via PI3K/AKT/NF-κB axis. Clin Transl Oncol. 2023;25(5):1402–12.
Liu Y, Wang D, Li Z, Li X, Jin M, Jia N, et al. Pan-cancer analysis on the role of PIK3R1 and PIK3R2 in human tumors. Sci Rep. 2022;12(1):5924.
Funding
This research was supported by the National Science Foundation of China [grant number 31870974].
Author information
Authors and Affiliations
Contributions
Conceptualization, JL; data curation, JL, NN and RM; formal analysis, JL and SZ; funding acquisition, GL and YL; investigation, JL; methodology, JL and QD; software, JL; supervision, GL; validation, JL and SC; visualization, JL; writing—original draft, JL and GL; writing—review and editing, GL and YL.
Corresponding authors
Ethics declarations
Conflict of interest
The authors assert that there are no conflicting interests to disclose.
Ethical standards
The present study followed the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendmets.
Informed consent
For this type of study, formal consent is not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12094_2023_3336_MOESM1_ESM.docx
Supplementary data to this article can be found online. Supplementary Table S1. The coefficients assessed by multivariate Cox regression. Supplementary Table S2. Correlations between the expression of NOTCH3 and PIK3R2 and clinical characteristics in melanoma patients. Supplementary Figure S1. Univariate Cox regression, OS, and ROC analysis of melanoma patients in the GSE65904 or GSE22155 datasets. (A)OS analysis of melanoma patients in the GSE22155 dataset. (B)ROC analysis of melanoma patients in the GSE22155 dataset. (C)Univariate Cox regression of melanoma patients in the GSE65904 dataset. (DOCX 266 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, J., Ma, R., Chen, S. et al. Anoikis patterns via machine learning strategy and experimental verification exhibit distinct prognostic and immune landscapes in melanoma. Clin Transl Oncol 26, 1170–1186 (2024). https://doi.org/10.1007/s12094-023-03336-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03336-w