Abstract
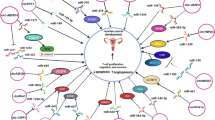
Ovarian cancer (OC) has the highest mortality rate among female reproductive system tumours, with limited efficacy of traditional treatments and 5-year survival rates that rarely exceed 40%. Circular RNA (circRNA) is a stable endogenous circular RNA that typically regulates protein expression by binding to downstream miRNA. It has been demonstrated that circRNAs play an important role in the proliferation, migration, and glucose metabolism (such as the Warburg effect) of OC and can regulate the expression of glucose metabolism-related proteins such as GLUT1 and HK2, promoting anaerobic glycolysis of cancer cells, increasing glucose uptake and ATP production, and affecting energy supply and biosynthetic substances to support tumour growth and invasion. This review summarises the formation and characteristics of circRNAs and focuses on their role in regulating glucose metabolism in OC cells and their potential therapeutic value, providing insights for identifying new therapeutic targets.


Similar content being viewed by others
Data availability
Not applicable.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Wang Z, Guo E, Yang B, Xiao R, Lu F, You L, et al. Trends and age-period-cohort effects on mortality of the three major gynecologic cancers in China from 1990 to 2019: cervical, ovarian and uterine cancer. Gynecol Oncol. 2021;163(2):358–63.
Armstrong DK, Alvarez RD, Backes FJ, Bakkum-Gamez JN, Barroilhet L, Behbakht K, et al. NCCN guidelines® insights: Ovarian cancer version 3.2022. J Natl Compr Canc Netw. 2022;20(9):972–80.
Morand S, Devanaboyina M, Staats H, Stanbery L, Nemunaitis J. Ovarian cancer immunotherapy and personalized medicine. Int J Mol Sci. 2021;22(12):6532.
O’Malley DM. New therapies for ovarian cancer. J Natl Compr Canc Netw. 2019;17(5.5):619–21.
Emmings E, Mullany S, Chang Z, Landen CN Jr, Linder S, Bazzaro M. Targeting mitochondria for treatment of chemoresistant ovarian cancer. Int J Mol Sci. 2019;20(1):229.
Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–37.
Fukushi A, Kim HD, Chang YC, Kim CH. Revisited metabolic control and reprogramming cancers by means of the warburg effect in tumor cells. Int J Mol Sci. 2022;23(17):10037.
Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8(6):519–30.
Vaupel P, Schmidberger H, Mayer A. The Warburg effect: essential part of metabolic reprogramming and central contributor to cancer progression. Int J Radiat Biol. 2019;95(7):912–9.
Icard P, Shulman S, Farhat D, Steyaert JM, Alifano M, Lincet H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist Updat. 2018;38:1–11.
Apostolova P, Pearce EL. Lactic acid and lactate: revisiting the physiological roles in the tumor microenvironment. Trends Immunol. 2022;43(12):969–77.
Chelakkot C, Chelakkot VS, Shin Y, Song K. Modulating glycolysis to improve cancer therapy. Int J Mol Sci. 2023;24(3):2606.
Ohshima K, Morii E. Metabolic reprogramming of cancer cells during tumor progression and metastasis. Metabolites. 2021;11(1):28.
Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21(10):669–80.
Abdel-Wahab AF, Mahmoud W, Al-Harizy RM. Targeting glucose metabolism to suppress cancer progression: prospective of anti-glycolytic cancer therapy. Pharmacol Res. 2019;150: 104511.
Tondo-Steele K, McLean K. The “Sweet Spot” of targeting tumor metabolism in ovarian cancers. Cancers (Basel). 2022;14(19):4696.
Tekade RK, Sun X. The Warburg effect and glucose-derived cancer theranostics. Drug Discov Today. 2017;22(11):1637–53.
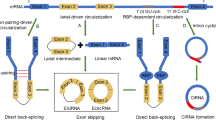
Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26(1):29.
Zhang C, Liu N. Noncoding RNAs in the glycolysis of ovarian cancer. Front Pharmacol. 2022;13: 855488.
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol. 2019;12(1):90.
Nava GM, Madrigal Perez LA. Metabolic profile of the Warburg effect as a tool for molecular prognosis and diagnosis of cancer. Expert Rev Mol Diagn. 2022;22(4):439–47.
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci. 2016;73(2):377–92.
Greene J, Segaran A, Lord S. Targeting OXPHOS and the electron transport chain in cancer; molecular and therapeutic implications. Semin Cancer Biol. 2022;86(Pt 2):851–9.
Morita M, Sato T, Nomura M, Sakamoto Y, Inoue Y, Tanaka R, et al. PKM1 Confers metabolic advantages and promotes cell-autonomous tumor cell growth. Cancer Cell. 2018;33(3):355-367.e7.
Israelsen WJ, Vander Heiden MG. Pyruvate kinase: function, regulation and role in cancer. Semin Cell Dev Biol. 2015;43(43):51.
İlhan M. Non-metabolic functions of pyruvate kinase M2: PKM2 in tumorigenesis and therapy resistance. Neoplasma. 2022;69(4):747–54.
Eniafe J, Jiang S. The functional roles of TCA cycle metabolites in cancer. Oncogene. 2021;40(19):3351–63.
Dier U, Shin DH, Hemachandra LP, Uusitalo LM, Hempel N. Bioenergetic analysis of ovarian cancer cell lines: profiling of histological subtypes and identification of a mitochondria-defective cell line. PLoS ONE. 2014;9(5): e98479.
Li N, Zhan X, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. 2018;150(2):343–54.
Anderson AS, Roberts PC, Frisard MI, McMillan RP, Brown TJ, Lawless MH, et al. Metabolic changes during ovarian cancer progression as targets for sphingosine treatment. Exp Cell Res. 2013;319(10):1431–42.
Xintaropoulou C, Ward C, Wise A, Queckborner S, Turnbull A, Michie CO, et al. Expression of glycolytic enzymes in ovarian cancers and evaluation of the glycolytic pathway as a strategy for ovarian cancer treatment. BMC Cancer. 2018;18(1):636.
Ma Y, Wang W, Idowu MO, Oh U, Wang XY, Temkin SM, et al. Ovarian cancer relies on glucose transporter 1 to fuel glycolysis and growth: anti-tumor activity of BAY-876. Cancers (Basel). 2018;11(1):33.
Baczewska M, Supruniuk E, Bojczuk K, Guzik P, Milewska P, Konończuk K, et al. Energy Substrate transporters in high-grade ovarian cancer: gene expression and clinical implications. Int J Mol Sci. 2022;23(16):8968.
Ciscato F, Ferrone L, Masgras I, Laquatra C, Rasola A. Hexokinase 2 in cancer: A prima donna playing multiple characters. Int J Mol Sci. 2021;22(9):4716.
Tian X, Liu D, Zuo X, Sun X, Wu M, Li X, et al. Hexokinase 2 promoted cell motility and proliferation by activating Akt1/p-Akt1 in human ovarian cancer cells. J Ovarian Res. 2022;15(1):92.
Suh DH, Kim MA, Kim H, Kim MK, Kim HS, Chung HH, et al. Association of overexpression of hexokinase II with chemoresistance in epithelial ovarian cancer. Clin Exp Med. 2014;14(3):345–53.
Garcia SN, Guedes RC, Marques MM. Unlocking the potential of HK2 in cancer metabolism and therapeutics. Curr Med Chem. 2019;26(41):7285–322.
Feng J, Li J, Wu L, Yu Q, Ji J, Wu J, et al. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39(1):126.
Lincet H, Kafara P, Giffard F, Abeilard-Lemoisson E, Duval M, Louis MH, et al. Inhibition of Mcl-1 expression by citrate enhances the effect of Bcl-xL inhibitors on human ovarian carcinoma cells. J Ovarian Res. 2013;6(1):72.
Kruspig B, Nilchian A, Orrenius S, Zhivotovsky B, Gogvadze V. Citrate kills tumor cells through activation of apical caspases. Cell Mol Life Sci. 2012;69(24):4229–37.
Wang X, Yin Y, Qian W, Peng C, Shen S, Wang T, et al. Citric acid of ovarian cancer metabolite induces pyroptosis via the caspase-4/TXNIP-NLRP3-GSDMD pathway in ovarian cancer. Faseb j. 2022;36(6): e22362.
Kopinski PK, Singh LN, Zhang S, Lott MT, Wallace DC. Mitochondrial DNA variation and cancer. Nat Rev Cancer. 2021;21(7):431–45.
Ju HQ, Lin JF, Tian T, Xie D, Xu RH. NADPH homeostasis in cancer: functions, mechanisms and therapeutic implications. Signal Transduct Target Ther. 2020;5(1):231.
Yang Y, Sauve AA. NAD(+) metabolism: Bioenergetics, signaling and manipulation for therapy. Biochim Biophys Acta. 2016;1864(12):1787–800.
Pramono AA, Rather GM, Herman H, Lestari K, Bertino JR. NAD- and NADPH-contributing enzymes as therapeutic targets in cancer: an overview. Biomolecules. 2020;10(3):358.
San-Millán I, Brooks GA. Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg effect. Carcinogenesis. 2017;38(2):119–33.
Zhang Y, Zhai Z, Duan J, Wang X, Zhong J, Wu L, et al. Lactate: the mediator of metabolism and immunosuppression. Front Endocrinol (Lausanne). 2022;13: 901495.
Stern R, Shuster S, Neudecker BA, Formby B. Lactate stimulates fibroblast expression of hyaluronan and CD44: the Warburg effect revisited. Exp Cell Res. 2002;276(1):24–31.
Ding J, Karp JE, Emadi A. Elevated lactate dehydrogenase (LDH) can be a marker of immune suppression in cancer: Interplay between hematologic and solid neoplastic clones and their microenvironments. Cancer Biomark. 2017;19(4):353–63.
Zhao J, Huang X, Xu Z, Dai J, He H, Zhu Y, et al. LDHA promotes tumor metastasis by facilitating epithelial-mesenchymal transition in renal cell carcinoma. Mol Med Rep. 2017;16(6):8335–44.
Liu M, Quek LE, Sultani G, Turner N. Epithelial-mesenchymal transition induction is associated with augmented glucose uptake and lactate production in pancreatic ductal adenocarcinoma. Cancer Metab. 2016;4:19.
Ghafouri-Fard S, Khoshbakht T, Hussen BM, Taheri M, Samsami M. Emerging role of circular RNAs in the pathogenesis of ovarian cancer. Cancer Cell Int. 2022;22(1):172.
Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19(3):188–206.
Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–40.
Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409.
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66.
Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44(3):1370–83.
Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–85.
Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–42.
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9): e1003777.
Nie H, Wang Y, Liao Z, Zhou J, Ou C. The function and mechanism of circular RNAs in gastrointestinal tumours. Cell Prolif. 2020;53(7): e12815.
Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–90.
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172.
Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. Faseb j. 1993;7(1):155–60.
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–30.
Li RC, Ke S, Meng FK, Lu J, Zou XJ, He ZG, et al. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9(8):838.
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417.
Luan W, Shi Y, Zhou Z, Xia Y, Wang J. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun. 2018;502(1):22–9.
Yu CY, Li TC, Wu YY, Yeh CH, Chiang W, Chuang CY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8(1):1149.
Sheng R, Li X, Wang Z, Wang X. Circular RNAs and their emerging roles as diagnostic and prognostic biomarkers in ovarian cancer. Cancer Lett. 2020;473:139–47.
Foruzandeh Z, Zeinali-Sehrig F, Nejati K, Rahmanpour D, Pashazadeh F, Seif F, et al. CircRNAs as potent biomarkers in ovarian cancer: a systematic scoping review. Cell Mol Biol Lett. 2021;26(1):41.
Liu F, Wu X, Zhu H, Wang F. Dysregulated expression of circular RNAs serve as diagnostic and prognostic markers in ovarian and cervical cancer: a PRISMA-compliant systematic review and meta-analysis. Medicine (Baltimore). 2021;100(39): e27352.
Luo Y, Gui R. Circulating exosomal circFoxp1 confers cisplatin resistance in epithelial ovarian cancer cells. J Gynecol Oncol. 2020;31(5): e75.
Chen Y, Ye X, Xia X, Lin X. Circular RNA ABCB10 correlates with advanced clinicopathological features and unfavorable survival, and promotes cell proliferation while reduces cell apoptosis in epithelial ovarian cancer. Cancer Biomark. 2019;26(2):151–61.
Liu N, Zhang J, Zhang LY, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(12):3713–8.
Ning L, Long B, Zhang W, Yu M, Wang S, Cao D, et al. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int J Oncol. 2018;53(6):2637–46.
Wang W, Wang J, Zhang X, Liu G. Serum circSETDB1 is a promising biomarker for predicting response to platinum-taxane-combined chemotherapy and relapse in high-grade serous ovarian cancer. Onco Targets Ther. 2019;12:7451–7.
Ning L, Lang J, Wu L. Plasma circN4BP2L2 is a promising novel diagnostic biomarker for epithelial ovarian cancer. BMC Cancer. 2022;22(1):6.
Qiu Y, Chen Y, Agbede O, Eshaghi E, Peng C. Circular RNAs in epithelial ovarian cancer: from biomarkers to therapeutic targets. Cancers (Basel). 2022;14(22):5711.
Chen S, Wu W, Li QH, Xie BM, Shen F, Du YP, et al. Circ-NOLC1 promotes epithelial ovarian cancer tumorigenesis and progression by binding ESRP1 and modulating CDK1 and RhoA expression. Cell Death Discov. 2021;7(1):22.
Xia B, Zhao Z, Wu Y, Wang Y, Zhao Y, Wang J. Circular RNA circTNPO3 regulates paclitaxel resistance of ovarian cancer cells by miR-1299/NEK2 signaling pathway. Mol Ther Nucleic Acids. 2020;21:780–91.
Yang C, Yuan W, Yang X, Li P, Wang J, Han J, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17(1):19.
Guo W, Zhang J, Zhang D, Cao S, Li G, Zhang S, et al. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget. 2017;8(29):48169–77.
Lin C, Xu X, Yang Q, Liang L, Qiao S. Circular RNA ITCH suppresses proliferation, invasion, and glycolysis of ovarian cancer cells by up-regulating CDH1 via sponging miR-106a. Cancer Cell Int. 2020;20:336.
Chen L, Zhang F, Sheng XG, Zhang SQ, Chen YT, Liu BW. MicroRNA-106a regulates phosphatase and tensin homologue expression and promotes the proliferation and invasion of ovarian cancer cells. Oncol Rep. 2016;36(4):2135–41.
Lai Y, Zhou B, Tan Q, Xu J, Wan T, Zhang L. LINC00116 enhances cervical cancer tumorigenesis through miR-106a/c-Jun pathway. J Cell Biochem. 2020;121(3):2247–57.
Zhao J, Klausen C, Qiu X, Cheng JC, Chang HM, Leung PC. Betacellulin induces Slug-mediated down-regulation of E-cadherin and cell migration in ovarian cancer cells. Oncotarget. 2016;7(20):28881–90.
Luo L, Gao YQ, Sun XF. Circular RNA ITCH suppresses proliferation and promotes apoptosis in human epithelial ovarian cancer cells by sponging miR-10a-α. Eur Rev Med Pharmacol Sci. 2018;22(23):8119–26.
Ye J, Wu D, Shen J, Wu P, Ni C, Chen J, et al. Enrichment of colorectal cancer stem cells through epithelial-mesenchymal transition via CDH1 knockdown. Mol Med Rep. 2012;6(3):507–12.
Bolaños JP, Almeida A, Moncada S. Glycolysis: a bioenergetic or a survival pathway? Trends Biochem Sci. 2010;35(3):145–9.
Qu D, Zou X, Liu Z. Propofol modulates glycolysis reprogramming of ovarian tumor via restraining circular RNA-zinc finger RNA-binding protein/microRNA-212-5p/superoxide dismutase 2 axis. Bioengineered. 2022;13(5):11881–92.
Liu H, Dilger JP, Lin J. Effects of local anesthetics on cancer cells. Pharmacol Ther. 2020;212: 107558.
Ren YL, Zhang W. Propofol promotes apoptosis of colorectal cancer cells via alleviating the suppression of lncRNA HOXA11-AS on miRNA let-7i. Biochem Cell Biol. 2020;98(2):90–8.
Yu X, Gao Y, Zhang F. Propofol inhibits pancreatic cancer proliferation and metastasis by up-regulating miR-328 and down-regulating ADAM8. Basic Clin Pharmacol Toxicol. 2019;125(3):271–8.
Sui H, Zhu C, Li Z, Yang J. Propofol suppresses gastric cancer tumorigenesis by modulating the circular RNA-PVT1/miR-195-5p/E26 oncogene homolog 1 axis. Oncol Rep. 2020;44(4):1736–46.
Wang P, Chen J, Mu LH, Du QH, Niu XH, Zhang MY. Propofol inhibits invasion and enhances paclitaxel- induced apoptosis in ovarian cancer cells through the suppression of the transcription factor slug. Eur Rev Med Pharmacol Sci. 2013;17(13):1722–9.
Olson SH, Carlson MD, Ostrer H, Harlap S, Stone A, Winters M, et al. Genetic variants in SOD2, MPO, and NQO1, and risk of ovarian cancer. Gynecol Oncol. 2004;93(3):615–20.
Bayer JL, Spitz DR, Christensen D, McCormick ML, Farley D, DeGeest K, et al. Biobehavioral and neuroendocrine correlates of antioxidant enzyme activity in ovarian carcinoma. Brain Behav Immun. 2015;50:58–62.
Yang H, Guo Y, Zhang Y, Wang D, Zhang G, Hou J, et al. Circ_MUC16 attenuates the effects of Propofol to promote the aggressive behaviors of ovarian cancer by mediating the miR-1182/S100B signaling pathway. BMC Anesthesiol. 2021;21(1):297.
Yang T, Cheng J, Yang Y, Qi W, Zhao Y, Long H, et al. S100B Mediates Stemness of Ovarian Cancer Stem-Like Cells Through Inhibiting p53. Stem Cells. 2017;35(2):325–36.
Yang T, Cheng J, You J, Yan B, Liu H, Li F. S100B promotes chemoresistance in ovarian cancer stem cells by regulating p53. Oncol Rep. 2018;40(3):1574–82.
Hou W, Zhang Y. Circ_0025033 promotes the progression of ovarian cancer by activating the expression of LSM4 via targeting miR-184. Pathol Res Pract. 2021;217: 153275.
Wu S, Le H. Dual roles of PKM2 in cancer metabolism. Acta Biochim Biophys Sin (Shanghai). 2013;45(1):27–35.
Pan Y, Xu T, Liu Y, Li W, Zhang W. Upregulated circular RNA circ_0025033 promotes papillary thyroid cancer cell proliferation and invasion via sponging miR-1231 and miR-1304. Biochem Biophys Res Commun. 2019;510(2):334–8.
Ye M, Hou H, Shen M, Dong S, Zhang T. Circular RNA circFOXM1 Plays a Role in Papillary Thyroid Carcinoma by Sponging miR-1179 and Regulating HMGB1 Expression. Mol Ther Nucleic Acids. 2020;19:741–50.
Xue R, Hua L, Xu W, Gao Y, Pang Y, Hao J. Derivation and validation of the potential core genes in pancreatic cancer for tumor-stroma crosstalk. Biomed Res Int. 2018;2018:4283673.
Wang J, Wu A, Yang B, Zhu X, Teng Y, Ai Z. Profiling and bioinformatics analyses reveal differential circular RNA expression in ovarian cancer. Gene. 2020;724: 144150.
Liu Y, He X, Chen Y, Cao D. Long non-coding RNA LINC00504 regulates the Warburg effect in ovarian cancer through inhibition of miR-1244. Mol Cell Biochem. 2020;464(1–2):39–50.
Xie, W., L.U. Liu, C. He, M. Zhao, R. Ni, Z. Zhang, et al., Circ_0002711 knockdown suppresses cell growth and aerobic glycolysis by modulating miR-1244/ROCK1 axis in ovarian cancer. J Biosci, 2021. 46.
Julian L, Olson MF. Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions. Small GTPases. 2014;5: e29846.
Ma X, Dang Y, Shao X, Chen X, Wu F, Li Y. Ubiquitination and long non-coding RNAs regulate actin cytoskeleton regulators in cancer progression. Int J Mol Sci. 2019;20(12):2997.
Reymond N, Im JH, Garg R, Cox S, Soyer M, Riou P, et al. RhoC and ROCKs regulate cancer cell interactions with endothelial cells. Mol Oncol. 2015;9(6):1043–55.
Zhang X, Zhang L, Du Y, Zheng H, Zhang P, Sun Y, et al. A novel FOXM1 isoform, FOXM1D, promotes epithelial-mesenchymal transition and metastasis through ROCKs activation in colorectal cancer. Oncogene. 2017;36(6):807–19.
Farooqi AA, Zahid R, Naureen H, Attar R, Gazouli M, Berardi R, et al. Regulation of ROCK1/2 by long non-coding RNAs and circular RNAs in different cancer types. Oncol Lett. 2022;23(5):159.
Vennin C, Rath N, Pajic M, Olson MF, Timpson P. Targeting ROCK activity to disrupt and prime pancreatic cancer for chemotherapy. Small GTPases. 2020;11(1):45–52.
Fu Y, Sun H. The molecular mechanism of circRHOBTB3 inhibits the proliferation and invasion of epithelial ovarian cancer by serving as the ceRNA of miR-23a-3p. J Ovarian Res. 2022;15(1):66.
Yalan, S., L. Yanfang, C. He, and T. Yujie, Circular RNA circRHOBTB3 inhibits ovarian cancer progression through PI3K/AKT signaling pathway. Panminerva Med, 2020.
Yang Q, Li F, He AT, Yang BB. Circular RNAs: Expression, localization, and therapeutic potentials. Mol Ther. 2021;29(5):1683–702.
Chen Y, Li C, Tan C, Liu X. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53(6):359–65.
Dahl M, Daugaard I, Andersen MS, Hansen TB, Grønbæk K, Kjems J, et al. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab Invest. 2018;98(12):1657–69.
Kristensen LS. Profiling of circRNAs using an enzyme-free digital counting method. Methods. 2021;196:11–6.
Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37(44):5829–42.
Lu M, Wu Y, Zeng B, Sun J, Li Y, Luo J, et al. CircEHMT1 inhibits metastatic potential of breast cancer cells by modulating miR-1233-3p/KLF4/MMP2 axis. Biochem Biophys Res Commun. 2020;526(2):306–13.
Funding
The Natural Science Foundation of Gansu Province of China (20JR10RA693) and by the Project of Administration of Traditional Chinese Medicine of Gansu Province of China (GZKG-2020-46) and Medical Innovation and Development Project of Lanzhou University (lzuyxcx-2022-200) and the intra-Hospital Funds of the First Hospital of Lanzhou University (No.ldyyyn2021-2).
Author information
Authors and Affiliations
Contributions
Conceptualisation, methodology and write the manuscript: YW, YY; software: XC; validation: YY. All the authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., Chen, X. & Yang, Y. CircRNA-regulated glucose metabolism in ovarian cancer: an emerging landscape for therapeutic intervention. Clin Transl Oncol 26, 584–596 (2024). https://doi.org/10.1007/s12094-023-03285-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03285-4