Abstract
Purpose
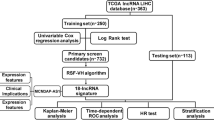
Long noncoding RNAs (lncRNAs) with abnormal expression are frequently seen in hepatocellular cancer patients (HCC). Previous studies have reported the correlation between lncRNA and prognosis processes of HCC patients. In this research, a graphical nomogram with lncRNAs signatures, T, M phases was developed using the rms R package to estimate the survival rates of HCC patients in year 1, 3, and 5.
Methods
To find the prognostic lncRNA and create the lncRNA signatures, univariate Cox survival analysis and multivariate Cox regression analysis were chosen. The rms R software package was used to build a graphical nomogram based on lncRNAs signatures to predict the survival rates in of HCC patients in 1, 3, and 5 years. Using “edgeR”, “DEseq” R packages to find the differentially expressed genes (DEGs).
Results
Firstly, a total of 5581 DEGs including 1526 lncRNAs and 3109 mRNAs were identified through bioinformatic analysis, of which 4 lncRNAs (LINC00578, RP11-298O21.2, RP11-383H13.1, RP11-440G9.1) were identified to be strongly related to the prognosis of liver cancer (P < 0.05). Moreover, we constructed a 4-lncRNAs signature by using the calculated regression coefficient. 4-lncRNAs signature is identified to significantly correlated with clinical and pathological characteristics (such as T stage, and death status of HCC patients).
Conclusions
A prognostic nomogram on the base of 4-lncRNAs markers was built, which is capable to accurately predict the 1-year, 3-year, and 5-year survival of HCC patients after the construction of the 4-lncRNAs signature linked with prognosis of HCC.






Similar content being viewed by others
References
Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, Global Burden of Disease Liver Cancer C, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3(12):1683–91.
European Association for the Study of the Liver. Electronic address EEE and European Association for the Study of the L EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182–236.
Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7(8):448–58.
Dhanasekaran R, Bandoh S, Roberts LR. Molecular pathogenesis of hepatocellular carcinoma and impact of therapeutic advances. F1000Research. 2016;5:879.
Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67(1):358–80.
Brown ZJ, Greten TF, Heinrich B. Adjuvant treatment of hepatocellular carcinoma: prospect of immunotherapy. Hepatology. 2019;70(4):1437–42.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7.
Qian X, Zhao J, Yeung PY, Zhang QC, Kwok CK. Revealing lncRNA structures and interactions by sequencing-based approaches. Trends Biochem Sci. 2019;44(1):33–52.
Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109(7):2093–100.
Charles Richard JL, Eichhorn PJA. Platforms for investigating LncRNA functions. SLAS Technol. 2018;23(6):493–506.
Lai Q, Spoletini G, Mennini G, Laureiro ZL, Tsilimigras DI, Pawlik TM, et al. Prognostic role of artificial intelligence among patients with hepatocellular cancer: a systematic review. World J Gastroenterol. 2020;26(42):6679–88.
Pinyol R, Montal R, Bassaganyas L, Sia D, Takayama T, Chau GY, et al. Molecular predictors of prevention of recurrence in HCC with sorafenib as adjuvant treatment and prognostic factors in the phase 3 STORM trial. Gut. 2019;68(6):1065–75.
Fu Q, Yang F, Xiang T, Huai G, Yang X, Wei L, et al. A novel microRNA signature predicts survival in liver hepatocellular carcinoma after hepatectomy. Sci Rep. 2018;8(1):7933.
Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67(3):603–18.
Zhang B, Li C, Sun Z. Long non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 are novel prognostic markers for pancreatic cancer. Am J Transl Res. 2018;10(8):2648–58.
Gao Q, Wang ZC, Duan M, Lin YH, Zhou XY, Worthley DL, et al. Cell culture system for analysis of genetic heterogeneity within hepatocellular carcinomas and response to pharmacologic agents. Gastroenterology. 2017;152(1):232-242.e4.
Munir R, Lisec J, Swinnen JV, Zaidi N. Lipid metabolism in cancer cells under metabolic stress. Br J Cancer. 2019;120(12):1090–8.
Lin Z, Lu D, Wei X, Wang J, Xu X. Heterogeneous responses in hepatocellular carcinoma: the achilles heel of immune checkpoint inhibitors. Am J Cancer Res. 2020;10(4):1085–102.
Li M, Hu J, Jin R, Cheng H, Chen H, Li L, et al. Effects of LRP1B regulated by HSF1 on lipid metabolism in hepatocellular carcinoma. J Hepatocell Carcinoma. 2020;7:361–76.
Jiang Y, Sun A, Zhao Y, Ying W, Sun H, Yang X, et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567(7747):257–61.
Sharpton SR, Schnabl B, Knight R, Loomba R. Current concepts, opportunities, and challenges of gut microbiome-based personalized medicine in nonalcoholic fatty liver disease. Cell Metab. 2021;33(1):21–32.
Nam SH, Bae MR, Roh JL, Gong G, Cho KJ, Choi SH, et al. A comparison of the 7th and 8th editions of the AJCC staging system in terms of predicting recurrence and survival in patients with papillary thyroid carcinoma. Oral Oncol. 2018;87:158–64.
Tellapuri S, Sutphin PD, Beg MS, Singal AG, Kalva SP. Staging systems of hepatocellular carcinoma: a review. Indian J Gastroenterol. 2018;37(6):481–91.
Bednarsch J, Czigany Z, Heise D, Joechle K, Luedde T, Heij L, et al. Prognostic evaluation of HCC patients undergoing surgical resection: an analysis of 8 different staging systems. Langenbecks Arch Surg. 2021;406(1):75–86.
Zhu X, Tian X, Yu C, Shen C, Yan T, Hong J, et al. A long non-coding RNA signature to improve prognosis prediction of gastric cancer. Mol Cancer. 2016;15(1):60.
Song P, Jiang B, Liu Z, Ding J, Liu S, Guan W. A three-lncRNA expression signature associated with the prognosis of gastric cancer patients. Cancer Med. 2017;6(6):1154–64.
Zheng S, Zheng D, Dong C, Jiang J, Xie J, Sun Y, et al. Development of a novel prognostic signature of long non-coding RNAs in lung adenocarcinoma. J Cancer Res Clin Oncol. 2017;143(9):1649–57.
Sui J, Yang S, Liu T, Wu W, Xu S, Yin L, et al. Molecular characterization of lung adenocarcinoma: a potential four-long noncoding RNA prognostic signature. J Cell Biochem. 2019;120(1):705–14.
Huang GW, Xue YJ, Wu ZY, Xu XE, Wu JY, Cao HH, et al. A three-lncRNA signature predicts overall survival and disease-free survival in patients with esophageal squamous cell carcinoma. BMC Cancer. 2018;18(1):147.
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–63.
Mahpour A, Mullen AC. Our emerging understanding of the roles of long non-coding RNAs in normal liver function, disease, and malignancy. JHEP Rep. 2020;3(1): 100177.
Zhao QJ, Zhang J, Xu L, Liu FF. Identification of a five-long non-coding RNA signature to improve the prognosis prediction for patients with hepatocellular carcinoma. World J Gastroenterol. 2018;24(30):3426–39.
Yan J, Zhou C, Guo K, Li Q, Wang Z. A novel seven-lncRNA signature for prognosis prediction in hepatocellular carcinoma. J Cell Biochem. 2019;120(1):213–23.
Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015;16(4):e173–80.
Funding
This study was supported by Social Development Mandatory Program of Qiqihar Science and Technology Bureau (No.SFGG-201951).
Author information
Authors and Affiliations
Contributions
QM and YD: are responsible for the conception or design of the work. WL, LL, ZH and SJ: contribute the acquisition, analysis, or interpretation of data for the work. All authors finally approved the manuscript version to be published. YD is the guarantor of the article.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by Ethical Committee of The Third Affiliated Hospital of Qiqihar Medical University and conducted in accordance with the ethical standards. (Approval number: 2018LL-11). All experiments were performed complying with the relevant regulations (Supplementary Table 2).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Informed consent
Subjects signed the informed consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12094_2023_3244_MOESM1_ESM.pdf
Supplementary Supplementary Figure 1. Survival status along with survival times of training group (a-d), testing group (e-h). file1 (PDF 301 KB)
12094_2023_3244_MOESM2_ESM.pdf
Supplementary Supplementary Figure 2. Clustering analysis of key prognostic lncRNAs by using different cut-of values file2 (PDF 115 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mo, Q., Li, W., Liu, L. et al. A nomogram based on 4-lncRNAs signature for improving prognostic prediction of hepatocellular carcinoma. Clin Transl Oncol 26, 375–388 (2024). https://doi.org/10.1007/s12094-023-03244-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03244-z