Abstract
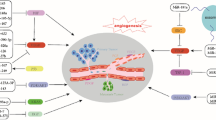
Colorectal cancer (CRC) is one of the most common cancers in the world. The incidence rate of cancer is high. The overall response to traditional treatment methods such as surgery, radiotherapy, and chemotherapy is not very satisfactory. Therefore, finding new therapeutic targets is very important for improving CRC treatment. In recent reports, the role of circRNAs in regulating colorectal angiogenesis has been gradually revealed. CircRNAs can indirectly act on angiogenesis pathways and regulate the expression of growth factors such as vascular endothelial growth factor (VEGF). CircRNAs are endogenous noncoding RNAs formed by pre-mRNAs through exon circular splicing. The covalent closed-loop structure makes these RNAs highly conserved and stable. CircRNAs have been found in human plasma, serum, urine, and other body fluids. Their highly conserved characteristics play important roles in many biological activities. CircRNAs can participate in the progression of many diseases by sponging miRNAs, interacting with proteins, and regulating transcription. Angiogenesis can provide nutrients and oxygen for tumour proliferation and metastasis. Angiogenesis is an important sign of the formation of the tumour microenvironment. Here, we will summarize the role of the latest circRNAs in the mechanism of angiogenesis in CRC and provide potential therapeutic targets for clinical treatment.


Similar content being viewed by others
Availability of data and materials
Not applicable.
References
Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Siegel RL, et al. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33.
Sung JJ, et al. Asia Pacific consensus recommendations for colorectal cancer screening. Gut. 2008;57(8):1166–76.
Dekker E, et al. Colorectal cancer. Lancet. 2019;394(10207):1467–80.
Tang Y, et al. MicroRNAs and angiogenesis: a new era for the management of colorectal cancer. Cancer Cell Int. 2021;21(1):221.
Seymour MT, et al. Different strategies of sequential and combination chemotherapy for patients with poor prognosis advanced colorectal cancer (MRC FOCUS): a randomised controlled trial. Lancet. 2007;370(9582):143–52.
Marisi G, et al. Circulating VEGF and eNOS variations as predictors of outcome in metastatic colorectal cancer patients receiving bevacizumab. Sci Rep. 2017;7(1):1293.
Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407(6801):249–57.
Carmeliet P, Jain RK. Principles and mechanisms of vessel normalization for cancer and other angiogenic diseases. Nat Rev Drug Discov. 2011;10(6):417–27.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Chen C, et al. The circular RNA 001971/miR-29c-3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. J Exp Clin Cancer Res. 2020;39(1):91.
Liu Y, Cao X. Characteristics and significance of the pre-metastatic niche. Cancer Cell. 2016;30(5):668–81.
Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20(4):409–26.
Bagnasco L, et al. Role of angiogenesis inhibitors in colorectal cancer: sensitive and insensitive tumors. Curr Cancer Drug Targets. 2012;12(4):303–15.
Widakowich C, et al. Review: side effects of approved molecular targeted therapies in solid cancers. Oncologist. 2007;12(12):1443–55.
Dai J, et al. CircRNA UBAP2 facilitates the progression of colorectal cancer by regulating miR-199a/VEGFA pathway. Eur Rev Med Pharmacol Sci. 2020;24(15):7963–71.
Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–40.
Ashwal-Fluss R, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66.
Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–11.
Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28(20):2233–47.
Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–57.
Li Z, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–64.
Huang C, et al. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32(9–10):639–44.
Ma Z, et al. circ5615 functions as a ceRNA to promote colorectal cancer progression by upregulating TNKS. Cell Death Dis. 2020;11(5):356.
Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8.
Abdelmohsen K, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–9.
Li A, et al. Circular RNA in colorectal cancer. J Cell Mol Med. 2021;25(8):3667–79.
Pamudurti NR, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9-21.e7.
Bak RO, Mikkelsen JG. miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA. 2014;5(3):317–33.
Shang A, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19(1):117.
Zhao X, Cai Y, Xu J. Circular RNAs: biogenesis, mechanism, and function in human cancers. Int J Mol Sci. 2019;20(16):3926.
Yang H, et al. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer. 2020;19(1):13.
Han K, et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol Cancer. 2020;19(1):60.
Zheng X, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18(1):47.
Pan Z, et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol Cancer. 2020;19(1):71.
Chen LY, et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation. Mol Cancer. 2020;19(1):164.
Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9(6):677–84.
Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. 2014;14(6):430–9.
Carmeliet P, et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394(6692):485–90.
Braunstein S, et al. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell. 2007;28(3):501–12.
Guo Y, et al. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer. 2021;20(1):93.
Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5.
Wu SY, Lan SH, Liu HS. Degradative autophagy selectively regulates CCND1 (cyclin D1) and MIR224, two oncogenic factors involved in hepatocellular carcinoma tumorigenesis. Autophagy. 2019;15(4):729–30.
Ramos-García P, et al. An update on the implications of cyclin D1 in oral carcinogenesis. Oral Dis. 2017;23(7):897–912.
Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16(9):398–406.
Bachmayr-Heyda A, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057.
Takahashi Y, et al. Expression of vascular endothelial growth factor and its receptor, KDR, correlates with vascularity, metastasis, and proliferation of human colon cancer. Cancer Res. 1995;55(18):3964–8.
Grothey A, Galanis E. Targeting angiogenesis: progress with anti-VEGF treatment with large molecules. Nat Rev Clin Oncol. 2009;6(9):507–18.
Jiang Z, et al. EIF4A3-induced circ_0084615 contributes to the progression of colorectal cancer via miR-599/ONECUT2 pathway. J Exp Clin Cancer Res. 2021;40(1):227.
Wang X, et al. MicroRNA-599 inhibits metastasis and epithelial-mesenchymal transition via targeting EIF5A2 in gastric cancer. Biomed Pharmacother. 2018;97:473–80.
Wang DP, et al. microRNA-599 promotes apoptosis and represses proliferation and epithelial-mesenchymal transition of papillary thyroid carcinoma cells via downregulation of Hey2-depentent Notch signaling pathway. J Cell Physiol. 2020;235(3):2492–505.
Sun Y, et al. MiR-429 inhibits cells growth and invasion and regulates EMT-related marker genes by targeting Onecut2 in colorectal carcinoma. Mol Cell Biochem. 2014;390(1–2):19–30.
Qi JH, et al. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med. 2003;9(4):407–15.
Zeng W, et al. CircFNDC3B sequestrates miR-937-5p to derepress TIMP3 and inhibit colorectal cancer progression. Mol Oncol. 2020;14(11):2960–84.
Jin L, et al. Circ_0030998 promotes tumor proliferation and angiogenesis by sponging miR-567 to regulate VEGFA in colorectal cancer. Cell Death Discov. 2021;7(1):160.
Zheng X, et al. Circ_0056618 promoted cell proliferation, migration and angiogenesis through sponging with miR-206 and upregulating CXCR4 and VEGF-A in colorectal cancer. Eur Rev Med Pharmacol Sci. 2020;24(8):4190–202.
Wu G, et al. Circ-RNF111 aggravates the malignancy of gastric cancer through miR-876-3p-dependent regulation of KLF12. World J Surg Oncol. 2021;19(1):259.
Kim SH, et al. Krüppel-like factor 12 promotes colorectal cancer growth through early growth response protein 1. PLoS ONE. 2016;11(7): e0159899.
Bai L, et al. Circular noncoding RNA circ_0007334 sequestrates miR-577 to derepress KLF12 and accelerate colorectal cancer progression. Anticancer Drugs. 2022;33(1):e409–22.
Funding
Chinese Foundation for Hepatitis Prevention and Control—‘Tian-qing-gan-bing’ Research Fund, No. TQGB20190165; College Students’ Innovation, Entrepreneurship and Excellence Program of Lanzhou University in 2022 (20220060024).
Author information
Authors and Affiliations
Contributions
Conceptualization, XG; Methodology, XC; Software, ZW; revised version of manuscript, CJ; Validation, ZW. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Guo, X., Chang, X., Wang, Z. et al. CircRNAs: promising factors for regulating angiogenesis in colorectal cancer. Clin Transl Oncol 24, 1673–1681 (2022). https://doi.org/10.1007/s12094-022-02829-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-022-02829-4