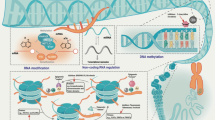
Abstract
Small B-cell lymphoma is the classification of B-cell chronic lymphoproliferative disorders that include chronic lymphocytic leukaemia/small lymphocytic lymphoma, follicular lymphoma, mantle cell lymphoma, marginal zone lymphoma, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia. The clinical presentation is somewhat heterogeneous, and its occurrence and development mechanisms are not yet precise and may involve epigenetic changes. Epigenetic alterations mainly include DNA methylation, histone modification, and non-coding RNA, which are essential for genetic detection, early diagnosis, and assessment of treatment resistance in small B-cell lymphoma. As chronic lymphocytic leukemia/small lymphocytic lymphoma has already been reported in the literature, this article focuses on small B-cell lymphomas such as follicular lymphoma, mantle cell lymphoma, marginal zone lymphoma, and Waldenstrom macroglobulinemia. It discusses recent developments in epigenetic research to diagnose and treat this group of lymphomas. This review provides new ideas for the treatment and prognosis assessment of small B-cell lymphoma by exploring the connection between small B-cell lymphoma and epigenetics.




Similar content being viewed by others
Availability of data and materials
Not applicable.
Abbreviations
- CLL:
-
Chronic lymphocytic leukaemia
- SLL:
-
Small lymphocytic lymphoma
- FL:
-
Follicular lymphoma
- MCL:
-
Mantle cell lymphoma
- MZL:
-
Marginal zone lymphoma
- LPL:
-
Lymphoplasmacytic lymphoma
- WM:
-
Waldenstrom macroglobulinemia
- MALT:
-
Mucosa-associated lymphoid tissue MZL
- NMZL:
-
Lymph node MZL
- SMZL:
-
Splenic MZL
- SAM:
-
S-adenosylmethionine
- DNMT:
-
Methyltransferase
- CGI:
-
CpG islands
- SHM:
-
Somatic hypermutation
- SIRT1:
-
Silent information regulator 1
- MBC:
-
Memory-like B cells
- PC:
-
Plasma cell
- PTM:
-
Post-translational modifications
- HAT:
-
Histone acetyltransferase
- HDAC:
-
Histone deacetylase
- HMT:
-
Histone methyltransferases
- PRC2:
-
Polyoma repressor complex 2
- H3K27:
-
Histone tris-lysine 27
- RNA-seq:
-
RNA sequencing
References
Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–90.
Nadeu F, Diaz-Navarro A, Delgado J, Puente XS, Campo E. Genomic and epigenomic alterations in chronic lymphocytic leukemia. Annu Rev Pathol. 2020;15:149–77.
Guièze R, Wu CJ. Genomic and epigenomic heterogeneity in chronic lymphocytic leukemia. Blood. 2015;126(4):445–53.
Martín-Subero JI, López-Otín C, Campo E. Genetic and epigenetic basis of chronic lymphocytic leukemia. Curr Opin Hematol. 2013;20(4):362–8.
Mansouri L, Wierzbinska JA, Plass C, Rosenquist R. Epigenetic deregulation in chronic lymphocytic leukemia: Clinical and biological impact. Semin Cancer Biol. 2018;51:1–11.
Li YY, Hu DZ, Tian C. Research progress on diagnosis and treatment of B cell chronic lymphoproliferative disease–review. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2018;26(4):1220–4.
Burger JA, Landau DA, Taylor-Weiner A, Bozic I, Zhang H, Sarosiek K, et al. Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition. Nat Commun. 2016;7:11589.
Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science (New York, NY). 2017;357(6348):eaal2380.
Memari F, Joneidi Z, Taheri B, Aval SF, Roointan A, Zarghami N. Epigenetics and Epi-miRNAs: potential markers/therapeutics in leukemia. Biomed Pharmacother. 2018;106:1668–77.
Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38.
Dan J, Chen T. Genetic studies on mammalian DNA methyltransferases. Adv Exp Med Biol. 2016;945:123–50.
Dario LS, Rosa MA, Mariela E, Roberto G, Caterina C. Chromatin remodeling agents for cancer therapy. Rev Recent Clin Trials. 2008;3(3):192–203.
Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochem Biophys Acta. 2007;1775(1):138–62.
Kretzmer H, Bernhart SH, Wang W, Haake A, Weniger MA, Bergmann AK, et al. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat Genet. 2015;47(11):1316–25.
Rossi D, Capello D, Gloghini A, Franceschetti S, Paulli M, Bhatia K, et al. Aberrant promoter methylation of multiple genes throughout the clinico-pathologic spectrum of B-cell neoplasia. Haematologica. 2004;89(2):154–64.
Hayslip J, Montero A. Tumor suppressor gene methylation in follicular lymphoma: a comprehensive review. Mol Cancer. 2006;5:44.
Giachelia M, Bozzoli V, D’Alò F, Tisi MC, Massini G, Maiolo E, et al. Quantification of DAPK1 promoter methylation in bone marrow and peripheral blood as a follicular lymphoma biomarker. J Mol Diagn. 2014;16(4):467–76.
Bennett LB, Schnabel JL, Kelchen JM, Taylor KH, Guo J, Arthur GL, et al. DNA hypermethylation accompanied by transcriptional repression in follicular lymphoma. Genes Chromosom Cancer. 2009;48(9):828–41.
Alhejaily A, Day AG, Feilotter HE, Baetz T, Lebrun DP. Inactivation of the CDKN2A tumor-suppressor gene by deletion or methylation is common at diagnosis in follicular lymphoma and associated with poor clinical outcome. Clin Cancer Res. 2014;20(6):1676–86.
Rogozin IB, Lada AG, Goncearenco A, Green MR, De S, Nudelman G, et al. Activation induced deaminase mutational signature overlaps with CpG methylation sites in follicular lymphoma and other cancers. Sci Rep. 2016;6:38133.
Frazzi R, Zanetti E, Pistoni M, Tamagnini I, Valli R, Braglia L, et al. Methylation changes of SIRT1, KLF4, DAPK1 and SPG20 in B-lymphocytes derived from follicular and diffuse large B-cell lymphoma. Leuk Res. 2017;57:89–96.
Martin-Subero JI, Ammerpohl O, Bibikova M, Wickham-Garcia E, Agirre X, Alvarez S, et al. A comprehensive microarray-based DNA methylation study of 367 hematological neoplasms. PLoS One. 2009;4(9):e6986.
Leshchenko VV, Kuo PY, Shaknovich R, Yang DT, Gellen T, Petrich A, et al. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010;116(7):1025–34.
Kirschbaum M, Frankel P, Popplewell L, Zain J, Delioukina M, Pullarkat V, et al. Phase II study of vorinostat for treatment of relapsed or refractory indolent non-Hodgkin’s lymphoma and mantle cell lymphoma. J Clin Oncol. 2011;29(9):1198–203.
Arribas AJ, Rinaldi A, Mensah AA, Kwee I, Cascione L, Robles EF, et al. DNA methylation profiling identifies two splenic marginal zone lymphoma subgroups with different clinical and genetic features. Blood. 2015;125(12):1922–31.
Hunter ZR, Xu L, Yang G, Tsakmaklis N, Vos JM, Liu X, et al. Transcriptome sequencing reveals a profile that corresponds to genomic variants in Waldenström macroglobulinemia. Blood. 2016;128(6):827–38.
Roos-Weil D, Giacopelli B, Armand M, Della-Valle V, Ghamlouch H, Decaudin C, et al. Identification of 2 DNA methylation subtypes of Waldenström macroglobulinemia with plasma and memory B-cell features. Blood. 2020;136(5):585–95.
Fan J, Krautkramer KA, Feldman JL, Denu JM. Metabolic regulation of histone post-translational modifications. ACS Chem Biol. 2015;10(1):95–108.
Schiza V, Molina-Serrano D, Kyriakou D, Hadjiantoniou A, Kirmizis A. N-alpha-terminal acetylation of histone H4 regulates arginine methylation and ribosomal DNA silencing. PLoS Genet. 2013;9(9):e1003805.
Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays. 1998;20(8):615–26.
Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189–95.
Jiang Y, Ortega-Molina A, Geng H, Ying HY, Hatzi K, Parsa S, et al. CREBBP inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Discov. 2017;7(1):38–53.
Desmots F, Roussel M, Pangault C, Llamas-Gutierrez F, Pastoret C, Guiheneuf E, et al. Pan-HDAC inhibitors restore PRDM1 response to IL21 in CREBBP-mutated follicular lymphoma. Clin Cancer Res. 2019;25(2):735–46.
Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277(28):25748–55.
Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8(11):890–6.
Vaswani RG, Gehling VS, Dakin LA, Cook AS, Nasveschuk CG, Duplessis M, et al. Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a potent and selective inhibitor of histone methyltransferase EZH2, suitable for phase I clinical trials for B-Cell lymphomas. J Med Chem. 2016;59(21):9928–41.
McCabe MT, Mohammad HP, Barbash O, Kruger RG. Targeting histone methylation in cancer. Cancer J (Sudbury, Mass). 2017;23(5):292–301.
Gil VS, Bhagat G, Howell L, Zhang J, Kim CH, Stengel S, et al. Deregulated expression of HDAC9 in B cells promotes development of lymphoproliferative disease and lymphoma in mice. Dis Model Mech. 2016;9(12):1483–95.
Lue JK, O’Connor OA, Bertoni F. Targeting pathogenic mechanisms in marginal zone lymphoma: from concepts and beyond. Ann Lymphoma. 2020;4:7.
Hatjiharissi E, Ngo H, Leontovich AA, Leleu X, Timm M, Melhem M, et al. Proteomic analysis of waldenstrom macroglobulinemia. Can Res. 2007;67(8):3777–84.
Roccaro AM, Sacco A, Jia X, Azab AK, Maiso P, Ngo HT, et al. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood. 2010;116(9):1506–14.
Kugel JF, Goodrich JA. Non-coding RNAs: key regulators of mammalian transcription. Trends Biochem Sci. 2012;37(4):144–51.
Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, et al. Transcriptional control of gene expression by microRNAs. Cell. 2010;140(1):111–22.
Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–87.
He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett. 2017;396:138–44.
Wang W, Corrigan-Cummins M, Hudson J, Maric I, Simakova O, Neelapu SS, et al. MicroRNA profiling of follicular lymphoma identifies microRNAs related to cell proliferation and tumor response. Haematologica. 2012;97(4):586–94.
Thompson MA, Edmonds MD, Liang S, McClintock-Treep S, Wang X, Li S, et al. miR-31 and miR-17-5p levels change during transformation of follicular lymphoma. Hum Pathol. 2016;50:118–26.
Zhao JJ, Lin J, Lwin T, Yang H, Guo J, Kong W, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115(13):2630–9.
Cai J, Liu X, Cheng J, Li Y, Huang X, Li Y, et al. MicroRNA-200 is commonly repressed in conjunctival MALT lymphoma, and targets cyclin E2. Graefe’s Arch Clin Exp Ophthalmol. 2012;250(4):523–31.
Zhu XW, Wang J, Zhu MX, Wang YF, Yang SY, Ke XY. MicroRNA-506 inhibits the proliferation and invasion of mantle cell lymphoma cells by targeting B7H3. Biochem Biophys Res Commun. 2019;508(4):1067–73.
Arribas AJ, Campos-Martín Y, Gómez-Abad C, Algara P, Sánchez-Beato M, Rodriguez-Pinilla MS, et al. Nodal marginal zone lymphoma: gene expression and miRNA profiling identify diagnostic markers and potential therapeutic targets. Blood. 2012;119(3):e9–21.
Bouteloup M, Verney A, Rachinel N, Callet-Bauchu E, Ffrench M, Coiffier B, et al. MicroRNA expression profile in splenic marginal zone lymphoma. Br J Haematol. 2012;156(2):279–81.
Arribas AJ, Gómez-Abad C, Sánchez-Beato M, Martinez N, Dilisio L, Casado F, et al. Splenic marginal zone lymphoma: comprehensive analysis of gene expression and miRNA profiling. Mod Pathol. 2013;26(7):889–901.
Zhou K, Yi S, Yu Z, Li Z, Wang Y, Zou D, et al. MicroRNA-223 expression is uniformly down-regulated in B cell lymphoproliferative disorders and is associated with poor survival in patients with chronic lymphocytic leukemia. Leuk Lymphoma. 2012;53(6):1155–61.
Pastore A, Gaiti F, Lu SX, Brand RM, Kulm S, Chaligne R, et al. Corrupted coordination of epigenetic modifications leads to diverging chromatin states and transcriptional heterogeneity in CLL. Nat Commun. 2019;10(1):1874.
Roccaro AM, Sacco A, Chen C, Runnels J, Leleu X, Azab F, et al. microRNA expression in the biology, prognosis, and therapy of Waldenström macroglobulinemia. Blood. 2009;113(18):4391–402.
Fulciniti M, Amodio N, Bandi RL, Cagnetta A, Samur MK, Acharya C, et al. miR-23b/SP1/c-myc forms a feed-forward loop supporting multiple myeloma cell growth. Blood Cancer J. 2016;6(1):e380.
Wang GG, Konze KD, Tao J. Polycomb genes, miRNA, and their deregulation in B-cell malignancies. Blood. 2015;125(8):1217–25.
Roisman A, Castellano G, Navarro A, Gonzalez-Farre B, Pérez-Galan P, Esteve-Codina A, et al. Differential expression of long non-coding RNAs are related to proliferation and histological diversity in follicular lymphomas. Br J Haematol. 2019;184(3):373–83.
Kienle D, Katzenberger T, Ott G, Saupe D, Benner A, Kohlhammer H, et al. Quantitative gene expression deregulation in mantle-cell lymphoma: correlation with clinical and biologic factors. J Clin Oncol. 2007;25(19):2770–7.
Garding A, Bhattacharya N, Claus R, Ruppel M, Tschuch C, Filarsky K, et al. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the In Cis downregulation of a gene cluster that targets NF-kB. PLoS Genet. 2013;9(4):e1003373.
Wang F, Cui D, Zhang Q, Shao Y, Zheng B, Chen L, et al. LncRNA00492 is required for marginal zone B-cell development. Immunology. 2022;165(1):88–98.
Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24(9):1609–20.
Eckschlager T, Plch J, Stiborova M, Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int J Mol Sci. 2017;18(7):1414.
Chen IC, Sethy B, Liou JP. Recent Update of HDAC Inhibitors in Lymphoma. Front Cell Dev Biol. 2020;8:576391.
Cogan JC, Liu Y, Amengual JE. Hypomethylating agents in lymphoma. Curr Treat Options Oncol. 2020;21(8):61.
Tiper IV, Webb TJ. Histone deacetylase inhibitors enhance CD1d-dependent NKT cell responses to lymphoma. Cancer Immunol Immunother. 2016;65(11):1411–21.
Bradshaw G, Sutherland HG, Haupt LM, Griffiths LR. Dysregulated MicroRNA expression profiles and potential cellular, circulating and polymorphic biomarkers in non-Hodgkin lymphoma. Genes. 2016;7(12):130.
Wu B, Li J, Wang H, Wu Q, Liu H. MiR-132-3p serves as a tumor suppressor in mantle cell lymphoma via directly targeting SOX11. Naunyn Schmiedebergs Arch Pharmacol. 2020;393(11):2197–208.
Gebauer N, Kuba J, Senft A, Schillert A, Bernard V, Thorns C. MicroRNA-150 Is up-regulated in extranodal marginal zone lymphoma of MALT type. Cancer Genomics Proteomics. 2014;11(1):51–6.
Di Lisio L, Martinez N, Montes-Moreno S, Piris-Villaespesa M, Sanchez-Beato M, Piris MA. The role of miRNAs in the pathogenesis and diagnosis of B-cell lymphomas. Blood. 2012;120(9):1782–90.
Ferrajoli A, Shanafelt TD, Ivan C, Shimizu M, Rabe KG, Nouraee N, et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. 2013;122(11):1891–9.
Musilova K, Mraz M. MicroRNAs in B-cell lymphomas: how a complex biology gets more complex. Leukemia. 2015;29(5):1004–17.
Saito Y, Suzuki H, Tsugawa H, Imaeda H, Matsuzaki J, Hirata K, et al. Overexpression of miR-142-5p and miR-155 in gastric mucosa-associated lymphoid tissue (MALT) lymphoma resistant to Helicobacter pylori eradication. PLoS One. 2012;7(11):e47396.
Lou X, Fu J, Zhao X, Zhuansun X, Rong C, Sun M, et al. MiR-7e-5p downregulation promotes transformation of low-grade follicular lymphoma to aggressive lymphoma by modulating an immunosuppressive stroma through the upregulation of FasL in M1 macrophages. J Exp Clin Cancer Res. 2020;39(1):237.
Husby S, Ralfkiaer U, Garde C, Zandi R, Ek S, Kolstad A, et al. miR-18b overexpression identifies mantle cell lymphoma patients with poor outcome and improves the MIPI-B prognosticator. Blood. 2015;125(17):2669–77.
He M, Gao L, Zhang S, Tao L, Wang J, Yang J, et al. Prognostic significance of miR-34a and its target proteins of FOXP1, p53, and BCL2 in gastric MALT lymphoma and DLBCL. Gastric Cancer. 2014;17(3):431–41.
Pan Y, Li H, Guo Y, Luo Y, Li H, Xu Y, et al. A pilot study of long noncoding RNA expression profiling by microarray in follicular lymphoma. Gene. 2016;577(2):132–9.
Tang X, Long Y, Xu L, Yan X. LncRNA MORT inhibits cancer cell proliferation and promotes apoptosis in mantle cell lymphoma by upregulating miRNA-16. Cancer Manag Res. 2020;12:2119–25.
Zhang J, Wei J, Wang Z, Feng Y, Wei Z, Hou X, et al. Transcriptome hallmarks in Helicobacter pylori infection influence gastric cancer and MALT lymphoma. Epigenomics. 2020;12(8):661–71.
Heuston EF, Lemon KT, Arceci RJ. The beginning of the road for non-coding RNAs in normal hematopoiesis and hematologic malignancies. Front Genet. 2011;2:94.
Ji T, Chen Q, Tao S, Shi Y, Chen Y, Shen L, et al. The research progress of circular RNAs in hematological malignancies. Hematology (Amsterdam, Netherlands). 2019;24(1):727–31.
Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. The critical role of RNA m(6)A methylation in cancer. Can Res. 2019;79(7):1285–92.
Zhang W, He X, Hu J, Yang P, Liu C, Wang J, et al. Dysregulation of N(6)-methyladenosine regulators predicts poor patient survival in mantle cell lymphoma. Oncol Lett. 2019;18(4):3682–90.
Wu G, Suo C, Yang Y, Shen S, Sun L, Li ST, et al. MYC promotes cancer progression by modulating m(6) A modifications to suppress target gene translation. EMBO Rep. 2021;22(3):e51519.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (82070175), Natural Science Foundation of Hunan Province (2021JJ30937), Scientific program of Health Commission of Hunan Province (2022030442723), and Changsha Municipal Natural Science Foundation (kq2014234).
Author information
Authors and Affiliations
Contributions
XR wrote the review. RZ, HZ, CY and ZW discussed the review. All authors proofread the review. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ruan, X., Zhang, R., Zhu, H. et al. Research progress on epigenetics of small B-cell lymphoma. Clin Transl Oncol 24, 1501–1514 (2022). https://doi.org/10.1007/s12094-022-02820-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-022-02820-z