Abstract
Purpose
Some patients with histologically confirmed primary mCRC and mutated RAS reported undetectable RAS mutant clones in plasma after receiving anti-VEGF treatment. The aim was to prospectively assess it with its potential therapeutic implications.
Methods
RAS mutant genes in solid biopsy (before first-line treatment: FOLFOX/CAPOX + bevacizumab) were compared in liquid biopsy (before second-line treatment: panitumumab + FOLFIRI), using Idylla™ system. Discordant results between solid/liquid biopsies were assessed by the next-generation sequencing (NGS) test (solid/liquid biopsies).
Results
Twenty-three patients were assessed (seven had RAS mutant discrepancies between solid/liquid biopsies). The NGS test confirmed that 3/23 (13%) patients had undetectable RAS mutant clones in liquid biopsy and 3/23 (13%) presented discrepancies in solid biopsy (Idylla™ system vs. NGS test).
Conclusion
Thirteen percentage of patients had undetectable RAS mutant clones in liquid biopsy after first-line treatment. However, some discrepancies between solid and liquid biopsies have been observed. These results suggest a need to improve accuracy of RAS analyses, especially in solid biopsies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Assessment of RAS mutations is crucial to guide treatment decisions in clinical practice. The determination must be performed in tumor tissue upon the diagnosis of metastatic disease either at the primary tumor or the metastatic disease [1].
Circulating cell-free tumor DNA (ctDNA) that originates from the currently present tumor has the same genetic and epigenetic alterations, which are related to tumor development, progression, and treatment resistance [2,3,4]. Moreover, ctDNA is more accurate than circulating tumor cells in respect of tumor burden and can be used as both a prognostic and diagnostic biomarker. It also acts as a predictor in the assessment of antineoplastic therapy through molecular analysis and mutation identification. TP53 and KRAS mutations, microsatellite instability or loss of heterozygosity, together with DNA hyper-methylation can be detected using ctDNA [5]. The analysis of ctDNA also so-called as a “liquid biopsy” has been proposed as an alternative to the invasive techniques for obtaining tumor samples. This liquid biopsy enables minimally invasive monitoring of tumor evolution over, and could provide current genetic information before initiate second-line treatment [2,3,4].
Some patients with primary mCRC and RAS mutation have reported undetectable RAS mutant clones in plasma after receiving anti-VEGF treatment. These patients were eligible for treatment with EGFR inhibitors and treated, achieving a clinical benefit. However, these results were reported in a small sample size, and the evidence of the clinical benefit with EGFR inhibitors was limited to one treatment [2].
The aim of this study was to prospectively assess the RAS genotype in patients with primary mCRC and mutated RAS in solid biopsy using Idylla™ system (before first-line treatment) and disease progression after first-line treatment with FOLFOX/CAPOX + bevacizumab. RAS genes in liquid biopsy (before second-line treatment: panitumumab + FOLFIRI) assessed with Idylla™ system were performed. Discordant results between solid biopsy and liquid biopsy (using Idylla™ system) were confirmed by the next-generation sequencing (NGS) test (in solid and liquid biopsies).
Materials and methods
Patients
Patients 18 years or older, with histologically confirmed primary mCRC, RAS mutant clones on primary tumor before first-line initiation, at least 1 lesion with ≥ 10 mm (according to RECIST criteria), ECOG performance status 0–2, who received FOLFOX/CAPOX + bevacizumab treatment (including patients who had discontinued oxaliplatin from the FOLFOX/CAPOX treatment due to neurotoxicity) and had a liquid biopsy prior to second-line initiation were included.
Study design
This study belonged to a phase II, multicenter, and single-arm clinical trial (2017-003242-25). It was performed in accordance with the Declaration of Helsinki, approved by the local ethics committees, and all patients gave their consent to participate.
Patients with RAS mutant mCRC in solid biopsy (before first-line treatment) were selected. The presence of RAS mutant clones was analyzed before second-line treatment in liquid biopsy.
Solid RAS mutations analyses
Analysis of RAS tissue point mutations before first-line treatment using Idylla™ system was performed in each center. The minor allele fraction (MAF) (Amplicon-sequence tissue) with this test is 5%.
Plasma ctDNA RAS mutations analyses
The analyses were done at the Complexo Hospitalario Universitario A Coruña (CHUAC) (Spain) using Idylla™ system (Biocartis, Mechelen, Belgium).
Differences in point mutations between solid biopsy (before first-line treatment) and liquid biopsy (before second-line treatment) with Idylla™ system were assessed using the NGS test (in solid and liquid biopsies) (VHIO Custom Amplicon-seq panel [6], at Vall d’Hebron Institute of Oncology (VHIO, Barcelona). The minimum variant allele frequency (MAF) was 3% for tissue samples and 1% for plasma samples.
Statistical analysis
Mean and standard deviation (SD) for continuous variables, and frequencies and percentages for categorical variables were obtained. Analyses were performed using SAS version 9.4.
Results
Twenty-three patients with primary mCRC and RAS mutation (on solid biopsy before first-line treatment) were screened in eight centers. Baseline characteristics are described in Table 1.
Among these screened patients, none met the selection criteria (19 patients do not meet the undetectable RAS mutant clones in liquid biopsy prior to second-line initiation, one did not have disease progression, one was on third-line treatment, one had interstitial pneumonitis, and one received not permitted medication).
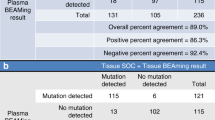
Although the study could not be performed, the results of RAS mutation analysis before first-line and second-line treatment, respectively, have reported new knowledge and learning about it (Fig. 1). RAS mutations were maintained before second-line treatment in 16 out of 23 (69.6%) patients (Idylla™ system) (Table 2). By contrast, some discrepancies between solid biopsy and liquid biopsy using Idylla™ system (seven out of 23 patients) were observed and were confirmed by the NGS test (Table 3). In four patients (1-003, 1-005, 1-007 and 4-001), RAS mutation in solid biopsy was not detected in liquid biopsy using Idylla™ system. Results in liquid biopsy using NGS test allowed to detect KRAS mutation in patient 1-003. Therefore, in three out of 23 patients (13.0%) (1-005, 1-007 and 4-001), undetectable ctDNA in liquid biopsy was verified by NGS test. In addition, in three patients (4-001, 3-002 and 2-002), some discrepancies in the RAS mutation genes (KRAS and NRAS genes) (Idylla™ system) were reported between solid and liquid biopsies. In these three patients, the results of NGS test confirmed some discrepancies vs. Idylla™ system, in solid biopsies: patient 4-001 reported NRAS mutation in solid biopsy using Idylla™ system when only BRAF mutation was confirmed by NGS test, patient 3-002 reported KRAS mutation in solid biopsy while it was confirmed by NGS test that this was a NRAS mutation. The patient 2-002 reported NRAS mutation in solid biopsy but it was confirmed by NGS test that this was a KRAS mutation. These analyses were repeated with Idylla™ system in the original tissue and corroborated that the initial results of solid biopsies with Idylla™ system had some mistakes.
Discussion
The genetic analysis of RAS genes of the screening patients that presented discordant results between solid and liquid biopsies (before first and second-line treatment, respectively) using Idylla™ system has allowed us to find some concerns that provide new evidence relevant in clinical practice.
The results of this study showed that most patients (16 patients, 70%) had concordance between solid (Gold Standard) and liquid biopsy results in RAS mutant clones using Idylla™ system. However, seven patients presented discordant results which were the reason to consider that the Gold Standard fails in the centers. Four of them had absence of any RAS mutations in plasma before initiation of second-line treatment. In three out of four, NGS test in plasma was also non-detectable. However, in the remaining patient (1-003), KRAS mutant could be detected in plasma with NGS test. This fact reinforces the higher sensitivity of NGS test vs. Idylla™ system. The patient’s sample that reported undetectable ctDNA in liquid biopsy with Idylla™ system presented 0.3% MAF.
Among the three patients with undetectable ctDNA, patient 1-007 was a patient who was still alive with low burden disease (2 cm lymphadenopathy: renal artery and terminal ileum) and patient 4-001 had metastasis in bone and paracolic gutter. By contrast, the patient 1-005 presented metastasis in lung and liver. The biological characteristic of the tumor is still poorly understood. Kagawa et al. [7] reported some discordances between plasma and tissues-based analyses related to the location of primary tumor, diameter, and number of metastatic lesions. Some studies reported a low proportion of undetectable RAS mutant clones (1.6–8.5%) in mCRC patients [8], while others found 45% (five out of 11) patients with RAS mutation not detected in plasma [2].
In the remaining three patients, some RAS mutations differences in solid biopsy using Idylla™ system and NGS test were observed. The genetic analyses were repeated a second time by the Idylla™ system in solid biopsy and support those obtained with the NGS test, which confirms that the previous results in Idylla™ system were most likely due to technical issues during routine testing. With the development of KRAS inhibitors, it is crucial to have an accurate result to know which codon and exon are mutated for a better therapeutic approach.
Colorectal cancer harbors a considerable heterogeneity, and the treatments imposed evolutionary pressure in selection of RAS mutations at progression of disease [9,10,11]. Genotyping cancer is mandatory in clinical practice to personalize the treatments. For example, recent evidence supports the use of the mismatch repair gene (MMR) testing for the implication of adjuvant therapy in patients with stage II colorectal cancer. The favorable prognosis of patients with stage II MSI-H colorectal cancer and the lack of benefit from adjuvant 5-fluorouracil-based therapy, indicate that these patients should avoid adjuvant chemotherapy. Therefore, testing for MMR status by MSI analysis or immunohistochemistry should be recommended in stage II colorectal cancer in patients who are candidates to adjuvant treatment is a consideration [12]. Therefore, improvement of good clinical practice to detect RAS mutation (especially using Idylla™ in solid biopsy), quality standard of care, monitorization, and the availability and the use of CEN technical documents and ISO standards for analytical procedures are needed.
This study has some limitations. This study was initially not designed to assess the genetic analysis of RAS genes in mCRC patients and the sample size was very small. The lack accessibility to liquid biopsies before first-line treatment is also an important limitation. To achieve an appropriate design, assessing RAS status in both solid and liquid biopsies should have been performed before first-line and before second-line treatment using both Idylla™ and NGS tests. In addition, the absence of detectable RAS mutations in plasma found before second-line treatment cannot certainly exclude that a RAS mutation might be present in the sample below the assay limit of detection. The sensitivity of the genetic analyses is relevant, and it is known that below 1% MAF, Idylla [13] has a reduced KRAS mutation detection in plasma.
Conclusion
Genotyping mCRC is crucial for personalized treatments. Our results showed some discrepancies between solid and liquid biopsies. Moreover, a lower percentage of undetectable RAS mutant clones compared with previous studies was observed. Therefore, there is a need for clinical improvement in the accuracy of the genotype analysis, especially in solid biopsies. As there is clonal selection, this type of approach should be implemented as part of the care of these patients, which will allow an adequate follow-up. In addition, this approach will not only be implemented in mCRC but also in localized disease.
References
Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27:1386–422.
Raimondi C, Nicolazzo C, Belardinilli F, Loreni F, Gradilone A, Mahdavian Y, et al. Transient disappearance of RAS mutant clones in plasma: a counterintuitive clinical use of EGFR inhibitors in RAS mutant metastatic colorectal cancer. Cancers. 2019;11:42.
Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–48.
Normanno N, Esposito Abate R, Lambiase M, Forgione L, Cardone C, Iannaccone A, et al. RAS testing of liquid biopsy correlates with the outcome of metastatic colorectal cancer patients treated with first-line FOLFIRI plus cetuximab in the CAPRI-GOIM trial. Ann Oncol. 2018;29:112–8.
Zarkavelis G, Boussios S, Papadaki A, Katsanos KH, Christodoulou DK, Pentheroudakis G. Current and future biomarkers in colorectal cancer. Ann Gastroenterol. 2017;30:613–21.
Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non–small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–2.
Kagawa Y, Elez E, Garcia-Foncillas J, Bando H, Taniguchi H, Vivancos A, et al. METABEAM study: combined analysis of concordance studies between liquid and tissue biopsies for RAS mutations in colorectal cancer patients with single metastatic sites. In: WCGIC Congress, vol 31, issue S3. 2020.
Henry J, Willis J, Parseghian CM, Raghav KPS, Johnson B, Dasari A, et al. NeoRAS: incidence of RAS reversion from RAS mutated to RAS wild type. JCO. 2020;38:180–180.
Oshima K, Khiabanian H, da Silva-Almeida AC, Tzoneva G, Abate F, Ambesi-Impiombato A, et al. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2016;113:11306–11.
Lu SX, Abdel-Wahab O. Genetic drivers of vulnerability and resistance in relapsed acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2016;113:11071–3.
Blank A, Roberts DE, Dawson H, Zlobec I, Lugli A. Tumor heterogeneity in primary colorectal cancer and corresponding metastases does the apple fall far from the tree? Front Med. 2018;5:234.
Boussios S, Ozturk MA, Moschetta M, Karathanasi A, Zakynthinakis-Kyriakou N, Katsanos KH, et al. The developing story of predictive biomarkers in colorectal cancer. J Pers Med. 2019;9:E12.
Vivancos A, Aranda E, Benavides M, Élez E, Gómez-España MA, Toledano M, et al. Comparison of the clinical sensitivity of the idylla platform and the OncoBEAM RAS CRC assay for KRAS mutation detection in liquid biopsy samples. Sci Rep. 2019;9:8976.
Acknowledgements
The authors wish to thank João Ramalho and Alfonso Troyano from Biocartis NV. Manuscript writing support was provided by Montse Sabaté, PhD from TFS Develop including: drafting the article, grammatical assistance, preparing references, and assembling tables. Medical writing was supported by Amgen S.A.
Funding
This work was supported by Amgen S.A. Amgen did not have any role in study design; collection, analysis, and interpretation of data; writing the report; and the decision to submit the report for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
FA has received compensation as advisor of Amgen S.A. The other authors declare no conflict of interest.
Ethical approval
The manuscript has not been published elsewhere and is not currently under consideration by another journal and all authors gave the consent for publication.
Informed consent
All patients gave their consent to participate in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fernández Montes, A., Élez, E., Vivancos, A. et al. Monitoring of RAS mutant clones in plasma of patients with RAS mutant metastatic colorectal cancer. Clin Transl Oncol 24, 1209–1214 (2022). https://doi.org/10.1007/s12094-021-02767-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-021-02767-7