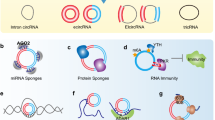
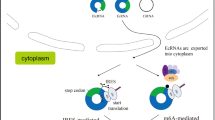
Abstract
Circular RNAs (circRNAs) have been considered a special class of non-coding RNAs without 5′ caps and 3′ tails which are covalently closed RNA molecules generated by back splicing of mRNA. For a long time, circRNAs have been considered to be directly involved in various biological processes as functional RNA. In recent years, a variety of circRNAs have been found to have translational functions, and the resultant peptides also play biological roles in the emergence and progression of human disease. The discovery of these circRNAs and their encoded peptides has enriched genomics, helped us to study the causes of diseases, and promoted the development of biotechnology. The purpose of this review is to summarize the research progress of the detection methods, translation initiation mechanism, as well as functional mechanism of peptides encoded by circRNAs, with the goal of providing the directions for the discovery of biomarkers for diagnosis, prognosis, and therapeutic targets for human disease.


Similar content being viewed by others
Abbreviations
- circRNA:
-
Circular RNA
- IRES:
-
Internal ribosome entry site
- ORF:
-
Open reading frame
- eIF:
-
Eukaryotic initiation factor
- m6A:
-
N6-methyladenosine
- YTHDF1/2/3:
-
YTH domain family protein 1/2/3
- GFP:
-
Green fluorescent protein
- Rluc:
-
Rellina luciferase
- Luc:
-
Luciferase
- PCNA:
-
Proliferating cell nuclear antigen
- USP28:
-
Ubiquitin-specific protease 28
- GSK3β:
-
Glycogen synthase kinase 3 beta
- AKT3:
-
Protein kinase B γ
- PDK1:
-
Pyruvate dehydrogenase kinase isozyme 1
- YAP1:
-
Yes-associated protein 1
- PAF1:
-
Polymerase-associated factor
- GPCR:
-
G-protein-coupled receptor
- Rspo:
-
R-spondins
- METTL3/14:
-
Methyltransferase-like 3/14
- FTO:
-
Fat mass and obesity-associated protein
- ALKBH5:
-
ALKB homolog 5
- IME:
-
Intron-mediated enhancement
References
Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21.
Chekulaeva M Rajewsky N. Roles of long noncoding RNAs and circular RNAs in Translation. Cold Spring Harb Perspect Biol. 2019;11:a032680.
Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–6.
Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57.
Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176:869–81.
Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806.
Zganiacz D, Milanowski R. Characteristics of circular ribonucleic acid molecules (circRNA). Postepy Biochem. 2017;63:221–32.
Chen N, Zhao G, Yan X, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19:218.
Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64.
Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8.
Conn VM, Hugouvieux V, Nayak A, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3:17053.
Fu L, Chen Q, Yao T, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget. 2017;8:43878–88.
Chen L, Zhang S, Wu J, et al. CircRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–61.
Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–64.
Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58.
Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–70.
Bi W, Huang J, Nie C, et al. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin Cancer Res. 2018;37:275.
Chang CY, Sarnow P. Initiation of protein synthesis by the eukaryotic. Transl Appar Circ RNAs Sci. 1995;268:415–7.
Xia X, Li X, Li F, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. 2019;18:131.
Zheng X, Chen L, Zhou Y, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18:47.
Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2019;37:1805–14.
Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in Repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304–15.
Zhang M, Zhao K, Xu X, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9:4475.
Legnini L, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37.
Zhao J, Lee EE, Kim J, et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun. 2019;10:2300.
Liang WC, Wong CW, Liang PP, et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84.
Banko JL, Klann E. Cap-dependent translation initiation and memory. Prog Brain Res. 2008;169:59–80.
Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45.
Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11.
Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–41.
Gu C, Zhou N, Wang Z, et al. CircGprc5a promoted bladder oncogenesis and metastasis through Gprc5a-targeting peptide. Mol Ther Nucleic Acids. 2018;13:633–41.
Zhi X, Zhang J, Cheng Z, et al. CircLgr4 drives colorectal tumorigenesis and invasion through Lgr4-targeting peptide. Int J Cancer. 2019. https://doi.org/10.1002/ijc.32549.
Imataka H, Olsen HS, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–25.
Morino S, Imataka H, Svitkin YV, et al. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap- dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol. 2000;20:468–77.
Liberman N, Gandin V, Svitkin YV, et al. DAP5 associates with eIF2β and eIF4AI to promote internal ribosome entry site driven translation. Nucleic Acids Res. 2015;43:3764–75.
Lamphear BJ, Kirchweger R, Skern T, et al. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–83.
Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–612.
Kearse MG, Wilusz JE. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev. 2017;31:1717–31.
Abe N, Hiroshima M, Maruyama H, et al. Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew Chem Int Ed Engl. 2013;52:7004–8.
Abe N, Matsumoto K, Nishihara M, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435.
Warner JR, Knopf PM, Rich A. A multiple ribosomal structure in protein synthesis. Proc Natl Acad Sci USA. 1963;49:122–9.
Chassé H, Boulben S, Costache V, et al. Analysis of translation using polysome profiling. Nucleic Acids Res. 2017;45:e15.
Ingolia NT, Ghaemmaghami S, Newman JR, et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–23.
Ingolia NT, Brar GA, Rouskin S, et al. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–50.
Dudekula DB, Panda AC, Grammatikakis I, et al. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42.
Wang J, Gribskov M. IRESpy: an XGBoost model for prediction of internal ribosome entry sites. BMC Bioinformatics. 2019;20:409.
Zhao J, Wu J, Xu T, et al. IRESfinder: identifying RNA internal ribosome entry site in eukaryotic cell using framed K-mer features. J Genet Genomics. 2018;45:403–6.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6.
Molinie B, Giallourakis CC. Genome-wide location analyses of N6-methyladenosine modifications (m6A-Seq). Methods Mol Biol. 2017;1562:45–53.
Plaza S, Menschaert G, Payre F. In search of lost small peptides. Annu Rev Cell Dev Biol. 2017;33:391–416.
Meng X, Chen Q, Zhang P, et al. CircPro: an integrated tool for the identification of circRNAs with protein-coding potential. Bioinformatics. 2017;33:3314–6.
Sun P, Li G. CircCode: a powerful tool for identifying circRNA coding ability. Front Genet. 2019;10:981.
Spriggs KA, Stoneley M, Bushell M, et al. Reprogramming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008;100:27–38.
Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–9.
Mo D, Li X, Raabe CA, et al. A universal approach to investigate circRNA protein coding function. Sci Rep. 2019;9:11684.
Funding
This work was supported by the National Natural Science Foundation of China (Grant number: no. 81602285 and 81872126), Nanjing Medical Science and technique Development Foundation (No. JQX17009).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participations or animals performed by any of the authors.
Informed consent
Informed consent is not required for this type of study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shi, Y., Jia, X. & Xu, J. The new function of circRNA: translation. Clin Transl Oncol 22, 2162–2169 (2020). https://doi.org/10.1007/s12094-020-02371-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-020-02371-1