Abstract
Objectives
Citron kinase (CIT-K), as a key Rho effector, functions to maintain proper structure of the midbody during cell mitosis. This study assessed CIT-K expression and its role in breast cancer cells.
Methods
Paraffin-embedded breast cancer and para-tumor tissues from 43 invasive breast cancer patients and 33 normal mammary glands were collected for immunohistochemistry. CIT-K expression knockdown was achieved using lentivirus carrying CIT-K shRNA in a wide range of breast cancer cell lines. Cells were then subjected to Western blot, qRT-PCR, cell proliferation, colony formation, transwell, and flow cytometric assays. The tumorigenicity of CIT-K knocked-down breast cancer cells was assessed using the nude mouse xenograft assay. Microarray analysis was performed to elucidate the underlying gene regulation after CIT-K silencing.
Results
CIT-K protein was overexpressed in breast cancer tissues, which is associated with advanced tumor stage, HER-2 expression and Ki-67 expression, whereas knockdown of CIT-K expression reduced breast cancer cell proliferation and colony formation, but promoted tumor cell apoptosis and cell-cycle arrest. Knockdown of CIT-K expression also inhibited breast cancer cell migration and invasion capacity. Moreover, CIT-K knockdown suppressed the tumorigenicity of breast cancer cells in nude mice. Molecularly, the expression of a variety of signaling genes, such as cyclin D1, EGFR, JAK1, TGF-α, PTK2, RAF1, RALB, SOS1, mTOR, and PTGS2, were altered after CIT-K knockdown.
Conclusions
This study demonstrated that CIT-K is associated with aggressive breast cancer behavior and targeting CIT-K may be a novel strategy for the future control of breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is a significant global health problem as one of the most frequently diagnosed malignancies in women [1], despite tremendous advances in early detection, prevention, treatment options, and the increased comprehension of its molecular and genetic basis. To date, breast cancer still remains the leading cause of cancer-related death in women due to tumor metastasis and resistance to therapies [2, 3]. Thus, there is an urgent need to identify and develop novel therapeutic targets and strategies to counter this devastating disease.
Cytokinesis is the most important and final step in cell mitosis, during which two daughter cells completely separate [4]. Failure to complete cytokinesis often leads to centrosome amplification and production of tetraploid cells and the latter being closely associated with tumorigenesis [5, 6]. Cytokinesis failure results in chromosomal instability that is associated with increased metastatic capacity and cancer drug-resistant properties [7, 8]. Many cytokinesis regulators, such as PLK1 and Aurora kinases, have been shown to play a crucial role in cancer development and progression, the targeting of which may have potentially therapeutic value against human cancers [9,10,11]. For example, small GTPases of the Rho family and their downstream effectors act as key regulators in various cell processes, including cell migration, adhesion, and cytokinesis [12, 13]. As a Rho effector conserved from insects to mammals, serine/threonine kinase citron (CIT-K) is mainly expressed in proliferating cells and primarily functions during the late stages of cytokinesis [14, 15]. Accumulating evidence suggests that CIT-K is important in the maintenance of midbody structure during cell mitosis and regulates the final abscission of the two daughter cells [16,17,18]. Studies have shown that CIT-K knockout mice display dramatic defective neurogenesis and testicular impairment due to dysregulated cytokinesis and massive apoptosis [14, 19]. However, the influence of CIT-K on human cancer development and progression is not yet well-defined [20]. A previous study showed that inhibition of CIT-K expression in hepatocellular carcinoma cells significantly inhibited tumor cell proliferation in vitro and tumorigenicity in vivo [21]. CIT-K was reported overexpressed in ovarian cancer tissue and its concentration is strongly associated with the therapeutic outcome of patients [22]. Another paper reported that CIT-K acted at the top of the hierarchy by controlling midbody formation and that elevated levels of CIT-K may lead to dysregulation of cytokinesis, which in-turn may facilitate the malignant transformation of cells [17]. Furthermore, a more recent study observed that knockdown of CIT-K expression dramatically decreased proliferation of breast, cervical, and colorectal cancer cells [23]. At the gene level, CIT-K knockdown-induced cytokinesis failure-triggered tumor cells to undergo apoptosis, which may occur through a p53-independent mechanism, especially in polyploid and actively dividing cancer cells [23]. Thus, in this study, we first assessed CIT-K expression in breast cancer tissues by comparing para-tumor tissues and normal mammary glands and the associated CIT-K expression with clinicopathological parameters from patients. We then investigated the effects of CIT-K expression knockdown on the regulation of breast cancer cell proliferation, colony formation, apoptosis, cell cycle distribution, and underlying molecular events in vitro and tumorigenicity in nude mice. We believe this study and its results provide useful information regarding the role of CIT-K in breast cancer.
Materials and methods
Cell lines and tissue specimens
Human breast cancer cell lines MDA-MB-231, MCF-7, T-47D, MCF-10A and MDA-MB-468 and a normal breast epithelial cell line MCF-10A were obtained from the Type Culture Collection of The Chinese Academy of Science (Shanghai, China) and grown in Gibco® Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS, Gibco, Grand Island NY, USA) at 37 °C in a 5% CO2 humidified incubator. In order to minimize the differences of cell cycle, cells in logarithmic growth phase were selected to when experiment involving cells were conducted.
A total of 115 paraffin-embedded human breast cancer and para-cancerous mammary gland tissues were obtained from 43 patients with infiltrating ductal breast carcinoma that underwent treatment at The First Affiliated Hospital of Xi’an Jiaotong University (Xi’an, China). The median age of these patients at diagnosis was 49 years and their clinicopathological characteristics are summarized in Table 1. True normal mammary gland tissues of 33 patients with benign breast diseases were also obtained from the same hospital. This study was approved by the medical ethics committee of The First Affiliated Hospital of Xi’an Jiaotong University.
Immunohistochemistry
Paraffin sections were dewaxed in xylene and rehydrated in a graded series of ethanol solution. For immunohistochemistry, tissue sections were subjected to antigen retrieval in a pressure cooker in 0.01 mol/L sodium citrate buffer, pH 6.0 at 121 °C/20 psi for 3–5 min, before being incubated with 3% H2O2 in phosphate buffered saline (PBS) to block potentially endogenous peroxidase activity. Then the sections were further incubated with 10% goat serum for 1 h to block nonspecific binding sites. After that, the sections were incubated overnight at 4 °C with a primary antibody [1% bovine serum albumin (BSA) in PBS-Tween 20 (PBST)], followed by incubation with a secondary antibody for 1 h. The sections were then incubated with a streptavidin–biotin complex (SABC) at 37 °C for 30 min. The color reaction was developed in diaminobenzidine (DAB) solution for 2 min and the sections were counterstained in hematoxylin and mounted with Immuno-mount (Thermo Scientific, Waltham, MA, USA). The scoring criteria were defined as follows: the average percentage of positively stained cells for 0, < 5%; 1, 5%–25%; 2, 26%–50%; and 3, > 50%. A total of three randomly selected visual fields per section were counted and each case that had a total score of greater than or equal to 3 was considered positive, whereas a score < 3 was considered negative.
Small hairpin RNA-mediated CIT-K knockdown
CIT-K (Sequence ID: NM_007174) shRNA and negative control (NC) shRNA were designed and cloned into the hU6-MCS-CMV-EGFP lentiviral vector (CIT-K shRNA, 5′-CAGGCAAGATTGAGAACG-3′ and NC shRNA, 5′-TTCTCCGAACGTGTCACGT-3′). After DNA sequence confirmation, these plasmids were then co-transfected with pCMV-dR 8.91 and pCMV-MD 2.G vectors into human embryonic kidney 293T cells (obtained from Genechem Co., Shanghai, China) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) to generate lentiviral particles, according to the manufacturer’s instructions. After the multiplicity of infection (MOI) was assessed, these lentiviral particles were then used in breast cancer MDA-MB-231 and T-47D cells to knockdown CIT-K expression.
Cell counting assay
Lentivirus-infected MDA-MB-231 and T-47D cells were seeded into 96-well plates at a density of 2 × 103 cells/well and incubated in a humidified 5% CO2 atmosphere at 37 °C. The relative cell numbers on 1, 2, 3, 4, and 5 days were counted using the Cellomics ArrayScan VTI (Thermo, Waltham, USA), according to the user’s manual. The cell proliferation rate was expressed as mean ± SD of triplicate experiments and summarized as a percentage of control.
Colony formation assay
Lentivirus-infected MDA-MB-231 and T-47D cells were seeded in 6-well plates at a density of 8 × 102 cells/well and incubated in a humidified 5% CO2 atmosphere at 37 °C for 14 days, with a medium change every 3 days. Then, the cells were washed with PBS, fixed in 4% paraformaldehyde, stained with Giemsa stain, and reviewed and photographed under an inverted microscope (OLYMPUS, Shinjuku, Japan). Cell colonies with 50 cells or more were counted and the data were expressed as mean ± SD of triplicate experiments and summarized as a percentage of control.
Transwell tumor cell migration and invasion assay
The invasion capacity of breast cancer cells, with or without lentiviral infection, was assessed via the transwell assay. The transwell filters with 8.0 µm pore size from Millipore (Switzerland) were precoated with Matrigel (BD, Franklin Lakes, NJ, USA), and diluted in DMEM at a ratio of 1:8. For the assay, cells (1 × 105) in 200 µL of serum-free DMEM were seeded into the upper compartments of the chambers. Meanwhile, the lower compartments of the chambers were filled with 500 µL of DMEM containing 20% FBS. After 24-h culture, tumor cells remaining on the top surface were removed using a cotton swab, whereas tumor cells invaded through Matrigel to the lower surface of the filters were fixed in 4% paraformaldehyde, stained with 0.1% crystal violet, and counted under an inverted microscope. The number of invaded cells was expressed as the average number of cells counted in five randomly selected visual fields per filter. Furthermore, tumor cell migration ability was assessed in the same way as the invasion assay, but the filters were without any Matrigel pre-coating.
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
Total cellular RNA was isolated from breast cancer cells using Trizol (Invitrogen) and reversely transcribed into complementary DNA using the SuperScript II Reverse Transcriptase and the oligo (dT) primer (Thermo). Quantitative PCR was then performed using a Step One Plus Real-Time PCR System and the Power SYBR Master Mix (Takara Bio, Japan), following the manufacturer’s instructions. The primer sequences used in this study are detailed in Table 2. The relative level of mRNA expression was calculated using the 2−ΔΔCt method with the Ct values normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA, an internal control.
Western blotting
Western blotting was used to detect the CIT-K expression level in four different breast cancer cell lines (MDA-MB-231, MCF-7, T-47D, and MDA-MB-468) and a normal breast cell line MCF10-A. And it was also used to verify the microarray results. Cells were lysed in the radioimmunoprecipitation assay buffer containing protein inhibitors, and protein samples were added with the sodium dodecyl sulfate (SDS) sample buffer, resolved in 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gel, and transferred onto PVDF membranes (Roche, Mannheim, Germany) for 90 min at 200 mA. The membranes were blocked for 1 h in 5% nonfat milk solution in a Tris-buffered saline-Tween 20 (TBST). Thereafter, immunoblotting was performed with primary antibodies against CIT-K (1:1000, Abcam, Cambridge, MA, USA), cyclin D1 (1:200, Abcam), EGFR (1:10000, Abcam), PTGS2 (1:1000, Abcam), RALB (1:1000, Abcam), and TGF-alpha (1:1000, Abcam) in the blocking buffer containing 5% nonfat milk overnight at 4 °C. The next day, after incubation with the corresponding secondary antibody for 1 h at room temperature, the positive protein bands were visualized using the enhanced chemiluminescence (ECL) Western Blotting substrate (Amersham, Sweden).
Flow cytometry
The changed cell cycle distribution after knockdown of CIT-K expression was assayed using a flow cytometric cell cycle distribution assay. Briefly, cells were infected with lentivirus carrying CIT-K shRNA or NC shRNA for 3 days and then fixed in 70% ethanol at 4 °C before being re-suspended in PBS containing 50 µg/mL propidium iodide (PI) and 500 µg/mL RNase A. After incubation for 30 min in the dark, flow cytometric analysis was conducted using the BD FACS Calibur flow cytometer.
The relative cell apoptosis rate after knockdown of CIT-K expression was assayed using a flow cytometry-Annexin V-APC assay. Specifically, cells were infected with lentivirus carrying CIT-K shRNA or NC shRNA for 3 days and the ratio of apoptotic cells was measured using the Annexin V-APC Apoptosis Detection Kit (eBioscience, San Diego, CA, USA), according to the manufacturer’s instructions, and measured using the BD FACS Calibur flow cytometer.
Nude mouse subcutaneous tumor assay
This study was approved by the Animal Care and Use Committee of Xi’an Jiaotong University (Xi’an, China). For animal experiments, 5-week-old female NSG mice were purchased from Shanghai Slac Laboratory Animal Co. Ltd (Shanghai, China) and housed in ventilated microisolator cages. The mice were divided into two groups of 10 mice per group and injected subcutaneously in the second pair of mammary fat pads with 1 × 106 MDA-MB-231 cells (CIT-K knockdown or control) per injection in 100 µL PBS. Tumor sizes were determined with calipers every other day by measuring the length and width for 28 days. Tumor volumes were calculated according to the following formula: volume (mm3) = (length × width × width)/2. At the end of the experiments, mice were killed and tumor xenografts were removed and weighed.
Microarray gene profiling analysis
In this part of research, MDA-MB-231 cells were selected as an subject because it showed the highest expression level of CIT-K among the four breast cancer cell lines in Western blotting analysis. A total of 6 samples (3 for CIT-K-NC group and 3 for CIT-K-KD group) were collected for microarray. Total RNA was isolated from CIT-K knocked down or negative control MDA-MB-231 cells using Trizol (Invitrogen) and amplified into amplified RNA (aRNA), using the GeneChip Hybridization Wash and Stain Kit (Affymetrix, USA). This was used as a probe for microarray hybridization, i.e., aRNA probe (1.5 µg each) was hybridized to Affymetrix Microarray 901253, according to the manufacturer’s instructions, and data analysis was performed using R-project 3.1.0, affy package. The clustering method was used to measure the reproducibility of microarray data and Pearson correlation analysis was further conducted to validate the reliability of result. Differential gene expression was reached when the level of expression was changed twofold and p < 0.05.
Gene function and pathway enrichment analysis was carried out using the KEGG database (http://www.genome.jp/kegg/pathway/html) and GSEA analysis was further performed with GSEA software (http://www.broadinstitute.org/gsea/index.jsp). The differential genes overlapped with KEGG_PATHWAYS_IN_CANCER dataset were submitted to Cytoscape 3.1.1 software (Leroy Hood, USA) to depict the gene regulatory network.
Statistical analysis
Statistical analyses were performed using the SPSS statistical package, version 16.0, for Windows (SPSS Inc., Chicago, IL, USA). Data were presented as the mean ± SD, and the association between the level of CIT-K expression and breast cancer was assessed using the χ2 test. A two-sided p value of < 0.05 was considered statistically significant.
Results
Overexpression of CIT-K protein is associated with advanced tumor stage, HER-2 expression and Ki-67 expression
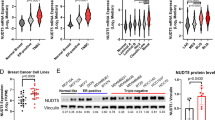
In this study, we first assessed CIT-K protein levels in different breast tissues and found that the number of CIT-K-positive cells was markedly higher in breast cancer samples than in the para-cancerous or normal breast tissues (Fig. 1a). The level of CIT-K mRNA in breast cancer tissues was also retrieved from The Cancer Genome Atlas (TCGA) with 1222 breast tissue samples. The data showed that compared with normal breast tissues, breast CIT-K was overexpressed in breast cancer (Fig. 1b). Furthermore, expression of CIT-K protein was associated with advanced tumor pathological grade (p < 0.05), HER-2 positivity (p < 0.05) and Ki-67 expression level (p < 0.05; Table 3). Accordingly, the positive rate of Ki-67 protein in CIT-K positive samples was also higher than that in CIT-K negative samples (Figure S1E, p < 0.05). In addition, through analysis of TCGA data, we found that the expression of CIT-K gene was positively correlated with MKI67 gene and TP63 gene in normal breast tissue (Figure S1C and D), but positively correlated with MKI67 gene and negatively correlated with TP63 gene in tumor tissue (Figure SA and B). However, there were no significant associations observed between CIT-K expression and other clinicopathological parameters (Table 3). The global view of CIT-K expression was further determined in different stages and types of breast cancer tissues through TCGA analyses. Compared with stage 1, breast cancer specimens from stage 4 that is at highest risk were characterized by up-regulated CIT-K (Fig. 1c). Intriguingly, when breast cancer samples were further divided into 3 molecular subtypes, relatively lower levels of CIT-K were found in Luminal A/B and HER2 + subtypes while the basal-like subtype showed the highest CIT-K expression (p < 0.01, Fig. 1d). However, the expression of CIT-K in the cancer tissue of these three subtypes was much higher than that in normal tissues, respectively (p < 0.001).
CIT-K expression in breast cancer tissue samples and cell lines. a Immunohistochemical analysis of CIT-K protein in breast cancer tissue specimens compared to para-cancerous or normal breast tissues. The statistical results show the proportion of CIT-K-positive cells in each tissue type. b TCGA database analysis of CIT-K mRNA levels of normal breast and breast cancer tissue samples. c TCGA database analysis of CIT-K mRNA levels in different stages of breast cancer tissue samples. Number of samples used: Stage I—181, Stage II—619, Stage III—247 and Stage IV—20. d TCGA database analysis of CIT-K mRNA levels in various types of breast cancer tissue samples. Number of samples used: Luminal A/B—440, HER2+—63, basal-like—115. e Western blot analysis of CIT-K protein in four breast cancer cell lines and a normal breast MCF-10A cell line. *P < 0.05 using Student’s t test. Values are mean ± SD
With this in mind, the levels of CIT-K protein in MDA-MB-231, MCF-7, T-47D, and MDA-MB-468 breast cancer cell lines were determined. We found that levels of CIT-K expression in all four breast cancer cell lines were remarkably higher than in the MCF-10A normal breast cell line (Fig. 1e). These results confirmed CIT-K overexpression in breast cancer and the association of CIT-K expression with breast cancer progression.
Knockdown of CIT-K expression decreases breast cancer cell proliferation and colony formation
To test the potential oncogenic activity of CIT-K in breast cancer progression, we knocked down CIT-K expression in breast cancer cells using lentivirus carrying CIT-K shRNA. Compared with negative control (NC) shRNA, CIT-K-knocked down MDA-MB-231 and T-47D cells exhibited significantly retarded cell proliferation (Fig. 2a). CIT-K knockdown also effectively inhibited colony formation in breast cancer cell lines (Fig. 2b). These findings point to the role of CIT-K in breast cancer progression.
Knockdown of CIT-K expression decreases breast cancer cell proliferation and colony formation. a Cell counting assay. MDA-MB-231 and T-47D cells were grown and infected with lentivirus carrying CIT-K shRNA and NC shRNA for 2 days and subjected to Cellomics cell counting. Each value represents the mean ± SD of triplicated wells. b Colony formation assay
Knockdown of CIT-K expression induces breast cancer cell apoptosis and cell-cycle arrest
We next investigated the root of the reduced cell proliferation and colony formation after knockdown of CIT-K expression in the two breast cancer cell lines tested, MDA-MB-231 and T-47D. The data demonstrates that compared with untreated or NC shRNA-infected cells, the number of apoptotic cells was significantly higher in CIT-K-KD cells (Fig. 3a, p < 0.01). Considering the crucial role of CIT-K during cytokinesis, we next examined if the distribution of cell-cycle phases changed after knockdown of CIT-K expression. As expected, the number of cells in the S phase of the cell cycle had significantly decreased but the G1 phase increased, compared to the controls (Fig. 3b, p < 0.01). To investigate whether the high expression of CIT-K is the driver for breast cancer malignance or the passenger of breast cancer development, we further investigated the effect of CIT-K knockdown on normal breast cell line. In sharp contrast to breast cancer cell lines, CIT-K knockdown resulted in no significant apoptosis or cell-cycle arrest in MCF-10A cell line (Fig. 3a, b), indicating that CIT-K may serve as an ideal target for cancer treatment. Combined, these data indicate that knockdown of CIT-K expression in breast cancer cells in vitro results in cell-cycle arrest and apoptosis in vitro.
Knockdown of CIT-K expression induces apoptosis and cell-cycle arrest in breast cancer cells. a Flow cytometric apoptosis assay. The proportions of apoptotic cells in CIT-K-KD, CIT-K-NC, and untreated control breast cancer cells were assayed by flow cytometry 2 days post-infection. The statistical results show the proportion of Annexin V+ PI− cells, which indicate apoptosis, but not necrosis, in each group. b Flow cytometric cell cycle distribution assay
Knockdown of CIT-K expression reduces breast cancer cell migration and invasion capacity
We also investigated the effects of CIT-K knockdown on the regulation of breast cancer cell migration and invasion. The transwell migration assay showed that the number of migrated CIT-K-KD MDA-MB-231 breast cancer cells was remarkably lower than control cells (Fig. 4a). In T-47D cells, a tendency of decreasing CIT-K-KD cell count could be observed (Fig. 4a), although not statistically. Moreover, the transwell invasion assay revealed that both CIT-K-KD MDA-MB-231 and CIT-K-KD T-47D cells reduced the tumor cell invasion capacity when compared to control cells (Fig. 4b). Collectively, these results demonstrated that CIT-K-KD breast cancer cells were much less potent in terms of migration and invasion.
Knockdown of CIT-K expression reduces breast cancer cell migration and invasion. a Transwell migration assay. MDA-MB-231 and T-47D cells were grown and infected with lentivirus carrying CIT-K shRNA and NC shRNA for 2 days and subjected to the transwell migration assay. The graph shows the quantity of migrated cells in each group. b Transwell invasion assay. MDA-MB-231 and T-47D cells were grown and infected with lentivirus carrying CIT-K shRNA and NC shRNA for 2 days and subjected to transwell Matrigel invasion assays. The graph shows the quantity of invaded cells in each group
Knockdown of CIT-K expression suppresses tumorigenicity in a breast cancer cell nude mouse model
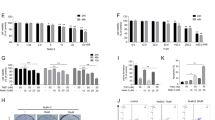
To extend our in vitro observations, we examined if knockdown of CIT-K expression affects breast cancer cell tumorigenic capacity in vivo. We subcutaneously injected CIT-K-KD MDA-MB-231 and NC shRNA cells into the flank of BALB/c nude mice. After allowing the tumor cells to grow in the mice for 28 days, the xenografts generated by CIT-K-KD cells were remarkably smaller in size than those of control groups (Fig. 5a). Furthermore, the growth curves and the xenograft wet weights showed statistical differences between the groups (p < 0.05, Fig. 5b, c). These data demonstrate that CIT-K not only accelerates proliferation of breast cancer cells in vitro, but also in vivo.
Knockdown of CIT-K expression suppresses breast cancer cell tumorigenicity in vivo. a Representative xenografts excised from the CIT-K-KD and negative control group injected nude mice and the associated tumor incidence in both groups, in terms of number of mice. b Tumor growth following subcutaneous injection of CIT-K-KD or control breast cancer cells. c Graph depicting the tumor weights of subcutaneous xenografts formed by CIT-K-KD or control breast cancer cells
Underlying mechanisms of CIT-K function
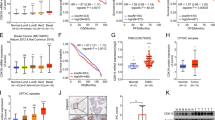
To investigate the molecular signaling regulated by CIT-K, we performed a microarray analysis of CIT-K knocked down and NC shRNA-infected MDA-MB-231 cells. We found a total of 266 up-regulated genes and 441 down-regulated genes between them (Supplemental Table 1). Through Gene Set Enrichment Analysis (GSEA), most of the differentially expressed genes were enriched in mitotic regulatory pathways, such as DNA replication and cell cycle (Fig. 6a), which is in accordance with the functions of CIT-K protein as a cytokinesis regulator. Interestingly, among all the candidate genes, 17 of them were involved in the gene set of the KEGG pathways in cancer. The potential gene regulatory network was depicted based on the known interaction information between CIT-K and these candidate genes (Fig. 6b). For verification, we examined expression of some of these genes in CIT-K-KD MDA-MB-231 cells using qRT-PCR. In agreement with the microarray result, CIT-K-KD cells exhibited marked reductions in the expression of cyclin D1, EGFR, JAK1, TGF-α, PTK2, and RAF1 mRNA, whereas knockdown of CIT-K expression dramatically increased levels of RALB, SOS1, mTOR, and PTGS2 mRNA (Fig. 6c, d, p < 0.01). We further assessed the expression of five genes that exhibited the most changes at the transcriptional level using a Western blot assay. Levels of cyclin D1, and TGF-α proteins were significantly down-regulated in CIT-K-KD MDA-MB-231 cells; level of EGFR has shown a down-regulated trend but did not reach a significant statistical difference; whereas PTGS2 expression was dramatically up-regulated (Fig. 6e). Similar changes were detected in the expression levels of these 5 proteins in CIT-K-KD T-47D cells. The only discrepancy observed was in RALB, which was up-regulated at the transcriptional level but was down-regulated at the protein level.
CIT-K knockdown regulated gene signaling pathways. a The GSEA analysis of molecular signaling pathways after CIT-K inhibition. b The potential gene regulatory network based on the known interaction information between CIT-K and the differentially expressed genes in the KEGG_PATHWAYS_IN_CANCER gene set. c qRT-PCR. Levels of cyclin D1, EGFR, JAK1, TGF-a, PTK2, and RAF1 mRNA were assayed using qRT-PCR in CIT-K-KD or negative control breast cancer cells. d qRT-PCR. Levels of RALB, SOS1, mTOR, and PTGS2 mRNA were assayed using qRT-PCR in CIT-K-KD or negative control breast cancer cells. e Western blot. Levels of cyclin D1, EGFR, TGF-a, PTGS2, and RALB proteins were analyzed by Western blot for both CIT-K-KD and negative control breast cancer cells
Discussion
In the current study, we demonstrated that CIT-K is an important regulator in the maintenance of breast cancer aggressiveness and tumorigenesis. Knockdown of CIT-K expression triggered tumor cell apoptosis and cell-cycle arrest, which may be caused by CIT-K knockdown-induced mitotic disorders and dysregulation of cancer-related signaling pathways.
Previous studies have revealed that CIT-K overexpression results in cytokinesis dysregulation and the production of polyploid cells, which is closely related to normal cell malignant transformation [24, 25]. In the current study, we observed high expression levels of CIT-K protein in breast cancer tissue specimens and cells compared with non-cancerous tissues, which is consistent with previous studies [26]. We also found that CIT-K expression was associated with advanced breast cancer stages, and HER-2 expression. Amplification of the HER-2 oncogene has been observed in 15–25% of all breast cancer cases [27] and HER-2-positive breast cancer has been shown to be more aggressive and have a higher recurrence rate and shorter overall survival (OS) than other subgroups [28].
It has been reported that Ki-67 expression in normal breast tissues is low (< 3%), and its positive rate shows an increasing trend in benign breast diseases, ductal carcinoma in situ and invasive breast cancer. Ki-67 is an important biological prognostic marker of breast cancer. High Ki-67 level often hints a higher recurrence rate and worse prognosis [29]. In this study, CIT-K expression may be positively correlated with Ki-67 expression, whether in the study of immunohistochemistry of cancer tissue or in the analysis of TCGA database. In addition, we found that the expression of CIT-K gene was positively correlated with TP63 gene in normal breast tissue, but negatively correlated with TP63 gene in tumor tissue. P63 is mainly expressed in the nuclei of myoepithelial cells of normal breast ducts and lobules and some special pathological type of breast cancer, such as metaplastic breast carcinomas, breast sarcomas and medullary breast cancer, while the positive rate of P63 in invasive breast cancer is extremely low [30]. Therefore, we speculate that the CIT-K is also higher in cancer cells with higher expression of Ki67, but the relationship between the expression of P63 and CIT-K needs to be further studied. The contradiction between CIT-K and P63 also indicates that there are many factors affecting the proliferation of cancer cells, and their genetic background is more complex than that of normal tissues. The effect of CIT-K overexpression on the proliferation of cancer cells is not the only or decisive factor. CIT-K expression rate might be an indirect marker of cancer cell proliferation, but should not be used as an absolute standard due to the complexity of proliferation-regulating mechanism of tumor cells.
Since CIT-K is a key Rho effector and mediates the formation of the midbody during cell mitosis, CIT-K expression may promote cell division and proliferation. We therefore, knocked down CIT-K expression in CIT-K-overexpressed invasive breast cancer cell lines using CIT-K shRNA. To increase transfection efficiency, we applied a lentivirus carrying CIT-K shRNA and NC shRNA to infect breast cancer cells for successful knockdown of CIT-K expression. We found that knockdown of CIT-K expression inhibited breast cancer cell proliferation in vitro and in vivo and reduced tumor cell migration and invasion capacity. In contrast, CIT-K knockdown accelerated breast cancer cell apoptosis and arrested tumor cells at the G1 phase of the cell cycle. A previous study, using a liver cancer cell model, showed that the CIT-K protein was usually expressed at the late S and/or the early G2 phase of the cell cycle following cyclin B1 expression [31]. Thus, loss of CIT-K expression was proven to result in cell-cycle arrest at the G2/M phase in both normal and cancerous hepatocytes [21, 31]. The reasons behind this inconsistency are unclear, but may be due to differences in cancer types or because of the existence of unique cell-cycle control mechanisms in breast cancer vs. liver cancer. Our current microarray data supported our findings, but further study is warranted to confirm and disclose how CIT-K regulates cell cycle progression and the underlying molecular events, perhaps in different cell models.
Furthermore, our current microarray data revealed a previously unknown connection between CIT-K and some key regulators in breast cancer cells. Among these altered genes, cyclin D1 stood out, as cyclin D1 plays a crucial role in mediating cell cycle and cancer progression. Members of the D-type cyclin family function as activators of cyclin-dependent kinase 4 (CDK4) and CDK6 to regulate the cell cycle transition from G1 to S phase [32]. Overexpression of cyclin D1 has been identified in a variety of human cancers and is associated with tumor metastasis and poor prognosis [33, 34]. Significant down-regulation of cyclin D1 after CIT-K depletion at both transcriptional and protein levels, suggests that cyclin D1 may serve as a downstream effector of CIT-K to bypass cell-cycle control and facilitate cancer cell proliferation. This novel finding may also explain why the majority of CIT-K knockdown breast cancer cells were arrested at the G1 phase of the cell cycle. Our current microarray results also revealed that there was no association between CIT-K knockdown and the classical Rho/ROCK pathway, indicating that CIT-K may exert its role through a Rho-independent pathway. The precise mechanism underpinning the CIT-K-cyclin D1 signaling network in breast cancer deserves further investigation.
However, there are still some principal limitations to the current study. Foremost, we did not provide data on gain-of-function experiments to further validate the function of CIT-K in breast cancer cells. Our findings would be decidedly strengthened if overexpression of CIT-K results in higher aggressiveness and tumorigenesis of breast cancer cells. Secondly, we did not investigate changes in binucleated and multinucleated cell populations after CIT-K knockdown and CIT-K dysregulation may cause the production of polyploid cells due to cytokinesis failure. Thirdly, we need to further study the expression and role of CIT-K in different molecular types of breast cancer cells or tissues. In conclusion, this study is the first to evaluate the effect of CIT-K knockdown on the regulation of breast cancer cell biological characteristics in vitro and in nude mice. The significant suppression of in vivo tumorigenicity after CIT-K depletion in breast cancer cells suggests the potential therapeutic value of CIT-K suppression as a target for drug development. However, since CIT-K knockdown also induced the up-regulation of a number of oncogenes identified by our current microarray analysis, the side effects of CIT-K-targeted cancer therapy warrant more in-depth investigation.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108.
Weigelt B, Peterse JL, van ‘t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602.
Núñez C, Capelo JL, Igrejas G, Alfonso A, Botana LM, Lodeiro C. An overview of the effective combination therapies for the treatment of breast cancer. Biomaterials. 2016;97:34–50.
Eggert US, Mitchison TJ, Field CM. Animal cytokinesis: from parts list to mechanisms. Annu Rev Biochem. 2016;75:543–66.
Sagona AP, Stenmark H. Cytokinesis and cancer. FEBS Let. 2010;584:2652–61.
Normand G, King RW. Understanding cytokinesis failure. Adv Exp Med Biol. 2010;676:27–55.
McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528–38.
Turajlic S, Swanton C. Metastasis as an evolutionary process. Science. 2016;352:169–75.
Ikezoe T, Yang J, Nishioka C, Takezaki Y, Tasaka T, Togitani K. A novel treatment strategy targeting polo-like kinase 1 in hematological malignancies. Cancer Res. 2009;23:1564–76.
Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, Maar S. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–84.
Li CC, Chu HY, Yang CW, Chou CK, Tsai TF. Aurora-A overexpression in mouse liver causes p53-dependent premitotic arrest during liver regeneration. Mol Cancer Res. 2009;7:678–88.
Ridley AJ. Rho family proteins: coordinating cell responses. Trends Cell Biol. 2001;11:471–7.
Thumkeo D, Watanabe S, Narumiya S. Physiological roles of Rho and Rho effectors in mammals. Eur J Cell Biol. 2013;92:303–15.
Di Cunto F, Imarisio S, Hirsch E, Broccoli V, Bulfone A, Migheli A. Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron. 2000;28:115–27.
Naim V, Imarisio S, Di Cunto F, Gatti M, Bonaccorsi S. Drosophila citron kinase is required for the final steps of cytokinesis. Mol Biol Cell. 2004;15:5053–63.
D’Avino PP, Capalbo L. Regulation of midbody formation and function by mitotic kinases. Semin Cell Dev Biol. 2016;53:57–63.
Bassi ZI, Audusseau M, Riparbelli MG, Callaini G, D’Avino PP. Citron kinase controls a molecular network required for midbody formation in cytokinesis. Proc Natl Acad Sci USA. 2013;110:9782–7.
Gai M, Camera P, Dema A, Bianchi F, Berto G, Scarpa E. Citron kinase controls abscission through RhoA and anillin. Mol Biol Cell. 2011;22:3768–78.
Cunto FD, Imarisio S, Camera P, Boitani C, Altruda F, Silengo L. Essential role of citron kinase in cytokinesis of spermatogenic precursors. J Cell Sci. 2002;115:4819–26.
Orgaz JL, Herraiz C, Sanz-Moreno V. Rho GTPases modulate malignant transformation of tumor cells. Small GTPases. 2014;5:e29019.
Fu Y, Huang J, Wang KS, Zhang X, Han ZG. RNA interference targeting CITRON can significantly inhibit the proliferation of hepatocellular carcinoma cells. Mol Biol Rep. 2011;38:693–702.
Ehrlichova M, Mohelnikova-Duchonova B, Hrdy J, Brynychova V, Mrhalova M, Kodet R. The association of taxane resistance genes with the clinical course of ovarian carcinoma. Genomics. 2013;102:96–101.
Kenzie C, D’Avino PP. Investigating cytokinesis failure as a strategy in cancer therapy. Oncotarget. 2016;7:87323–41.
Anastas SB, Mueller D, Semple-Rowland SL, Breunig JJ, Sarkisian MR. Failed cytokinesis of neural progenitors in citron kinase-deficient rats leads to multiciliated neurons. Cereb Cortex. 2011;21:338–44.
Serres MP, Kossatz U, Chi Y, Roberts JM, Malek NP, Besson A. p27(Kip1) controls cytokinesis via the regulation of citron kinase activation. J Clin Invest. 2012;122:844–58.
Brynychova V, Ehrlichova M, Hlavac V, Nemcova-Furstova V, Pecha V, Leva J. Genetic and functional analyses do not explain the association of high PRC1 expression with poor survival of breast carcinoma patients. Biomed Pharmacother. 2016;83:857–64.
Ross JS, Slodkowska EA, Symmans WF, Pusztai L, Ravdin PM, Hortobagyi GN. The HER-2 receptor and breast cancer: ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist. 2009;14:320–68.
Witton CJ, Reeves JR, Going JJ, Cooke TG, Bartlett JM. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J Pathol. 2003;200:290–7.
Denkert C, von Minckwitz G. Reply to Ki67 in breast cancer: a useful prognostic marker! Ann Oncol. 2014;25:542–3.
Di Franco S, Sala G, Todaro M. p63 role in breast cancer. Aging (Albany NY). 2016;8:2256–7.
Liu H, Di Cunto F, Imarisio S, Reid LM. Citron kinase is a cell cycle-dependent, nuclear protein required for G2/M transition of hepatocytes. J Biol Chem. 2003;278:2541–8.
Coleman ML, Marshall CJ. A family outing: small GTPases cyclin’ through G1. Nat Cell Biol. 2001;3:E250–1.
Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–72.
Qie S, Diehl JA. Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med (Berl). 2016;94:1313–26.
Acknowledgements
The authors would like to thank the Laboratory of Environmental and Disease Related Genes, Xi’an Jiaotong University for technical support during the study. This study was supported by funds from Basic Research Funds Project of Xi’an Jiaotong University (No. 10120121101). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All the authors declare that there is no conflict of interest in this work.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All procedures performed in studies involving animals were in accordance with the ethical standards of the Animal Care and Use Committee of Xi’an Jiaotong University (Xi’an, China).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
12094_2018_2003_MOESM1_ESM.jpg
The co-relationship of the expression of CIT-K, MKI67 and TP63 in breast cancer tissue samples and normal tissues in TCGA database. The expression of CIT-K was positive associated with cell proliferation marker MKI67 (A) but negative associated with TP63 (B) (n=1102) in breast carcinoma samples. In the compared normal tissues, the expression of CIT-K was positive associated with MKI67 (C) and TP63 (D) (n=113). (E) Immunohistochemical analysis of Ki-67 protein in breast cancer tissue specimens, and the CIT-K (+) patients showed a higher Ki-67expression level than CIT-K (-) patients. *P < 0.05 using Student’s t test (JPEG 7404 kb)
Rights and permissions
OpenAccess This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Meng, D., Yu, Q., Feng, L. et al. Citron kinase (CIT-K) promotes aggressiveness and tumorigenesis of breast cancer cells in vitro and in vivo: preliminary study of the underlying mechanism. Clin Transl Oncol 21, 910–923 (2019). https://doi.org/10.1007/s12094-018-02003-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-018-02003-9