Abstract
Analyze clinical samples collected and determine the etiology of viral pathogens and the dynamics of their spread. Acute respiratory viral infections remain one of the key health problems worldwide. They constitute etiologically independent diseases, with similar clinical infection manifestations and a single mechanism for the transmission of pathogens. 4712 nasopharyngeal swabs were collected from people before and during the COVID-19 pandemic with acute respiratory infections that tested negative for COVID-19 and were examined in this study. The collected samples were screened by a real-time polymerase chain reaction on a Rotor-Gene Q6 plex instrument. Statistical processing of the results, tabular, and graphical data were analyzed in the MS Excel. The largest number of the nasopharyngeal swabs were collected from children under 17 years of age (60.75%). In 702 samples (9.85%) pathogens of respiratory infections of non-influenza etiology were detected, including adenovirus, bocavirus, coronavirus, metapneumovirus, paramyxovirus types I–IV, respiratory syncytial virus, and rhinovirus. At the same time, both before and during the COVID-19 pandemic, different influenza virus variants co-circulation (A/H1N1, A/H3N2, and type B) were discovered, with a predominance of viruses with the antigenic formula A/H1N1. The results of the study indicate the need for continuous monitoring of the viral pathogens spread, which will expand the existing knowledge of the viral etiology of respiratory diseases and highlight the importance of viruses in the respiratory infections occurrence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute respiratory viral infections (ARVIs) have been affecting people since the beginning of human history and continue to be a serious health issue, leading to a significant health and economic damage. Most viral infections are zoonotic or vector-borne diseases caused by pathogens belonging to the families of Orthomyxoviridae, Paramyxoviridae, Picornaviridae, Coronaviridae, Adenoviridae, and Herpesviridae [1]. The most common infections of the upper respiratory tract account for 20–40% of outpatient and 12–35% of inpatient clinic attendance [2]. On average, there are 3–4 cases of the disease for every adult per year. Every year pathogens such as influenza, respiratory syncytial virus, adenovirus, parainfluenza virus, human coronavirus, human metapneumovirus, rhinovirus, enterovirus cause 30–40% of diseases leading to temporary disability among the population of the world and 60–80% of diseases resulting in loss of school hours among pupils. They also cause millions of deaths [3,4,5].
In recent times, coronaviruses, which were not previously classified as especially dangerous viral pathogens, especially a novel Severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) that emerged in 2019 [6], have become epidemiological significant.
Influenza viruses of the Orthomyxoviridae family occupy a dominant position among a large group of respiratory infections [7]. They include a wide range of human and animal pathogens [8,9,10,11,12,13]. The range of influenza virus epidemic strains is continually changing. In recent years, a rapid spread of influenza viruses of subtypes A/H1N1, A/H3N2, and type B is being observed in Kazakhstan as well as throughout the world [14, 15]. In addition, the extraordinary complexity of the epidemic situation is associated with the emergence of reassortant viruses.
Laboratory diagnostic procedures not only facilitate the identification of already known viruses, but also make it possible to detect previously unknown viruses. Thus, human metapneumovirus (hMpv) and human bocavirus (hBov) have been identified [16, 17]. The respiratory tract damage (and frequently a combined infection of the respiratory and gastrointestinal tracts) is caused by both deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) viruses. The same type of symptoms develops very frequently with various ARVIs, and, conversely, clinical manifestations may vary in diseases caused by a similar etiological agent [18, 19]. This requires a more active molecular biological study of various infections to possibly predict the severity of the disease and administer optimal treatment [20].
Therefore, the purpose of this study was to analyze clinical samples collected from people who attended healthcare facilities, determine the etiology of viral pathogens, and identify the dynamics of their spread.
Materials and Methods
Objects of Study
Nasopharyngeal swabs collected in 2018–2022 from people who attended healthcare facilities with acute respiratory diseases and showed a negative COVID-19 test result.
Sample Selection, Viral RNA Isolation and RT-PCR
The biomaterials (nasopharyngeal swabs) were collected in accordance with WHO recommendations [21]. The collected biosamples transported and stored in liquid nitrogen, while avoiding refreezing. The collected biomaterials were analyzed for the presence of the examined viruses using real-time polymerase chain reaction (RT-PCR) with hybridization-fluorescence detection on a Rotor-Gene Q6 plex device (QIAGEN, Germany). Extraction of nucleic acids and detection of viruses were performed with the RIBO-prep, AmpliSens ARVI-screen-FL, and AmpliSens® Influenza virus A/B-FL kits; the subtypes of influenza type A virus were identified with the AmpliSens® Influenza virus A-type-FL kit (Federal Budgetary Institution Central Research Institute for Epidemiology of Rospotrebnadzor, Russia) [22, 23].
Statistical Analysis
The software program MS Excel and the Graph Pad Prism 9 were used for statistical data processing, tabular, and graphic representation of the obtained results [24, 25]. Percentages were calculated for categorical variables such as levels of respiratory viruses. A chi-square test was used to assess the significance of intra-group differences for virus level by years. P ≤ 0.05 was considered statistically significant.
Results
In order to study the spread of pathogens causing ARVIs in 2018–2022, we examined a total of 4712 nasopharyngeal swabs from individuals of various ages (born between 1934–2022) with various respiratory diseases who had a negative SARS-CoV-2 test result (Fig. 1).
Most of the samples were collected from children under 17 years old inclusive, which represented 60.75% of the total number of samples. The largest people group of the working age patients (18–63 years) was represented by 22.71% of samples. The samples taken from people of retirement age accounted for about 16.54%.
The maximum number of samples (84.59%) was obtained from patients with ARVIs. The samples from patients diagnosed with acute respiratory disease, pneumonia, and bronchitis represented a smaller share (2.59% to 10.12%). Less than 1% of the samples were collected from patients with other disorders in the respiratory tract, such as tonsillitis, asthma, mucoviscidosis, etc.
Most of the samples were collected in 2018 and 2019 (2911 swabs) as compared to 2020–2022 (1,801 swabs). Number of samples collected during the 2020–2022 period decreased significantly compared to 2018–2019 due to the introduction of a special mode of operation in healthcare facilities associated with the COVID-19 pandemic (Fig. 2).
Samples were collected from healthcare facilities located in various regions of Kazakhstan, including the northern part of the country (Kostanay, Akmola, Pavlodar, and North Kazakhstan regions), the eastern part (East Kazakhstan Region), the southern part (Almaty, Zhambyl, South Kazakhstan, and Kazylorda regions), the western part (Mangystau, Atyrau, Aktobe, and West Kazakhstan Region), and the central area (Karaganda Region) (Fig. 3).
Most of the samples were collected from the southern part of Kazakhstan. This is explained by the restriction of movement around the country in accordance with the general anti-pandemic measures during 2020–2022.
As a result of RT-PCR testing, the genetic material for one of the detectable pathogens (i.e., adenovirus (hAdv), hBov, coronavirus (hCov), hMpv, paramyxovirus types I-IV (hPiv), respiratory syncytial virus (hRSv), rhinovirus (hRv), and influenza A and B viruses (hIv) detected in patients) was revealed in 1,847 samples (39.20% of the total number of the samples examined) (Table 1).
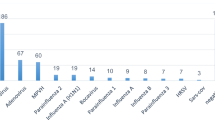
The viruses that cause respiratory infections of non-influenza etiology were detected in 702 samples, with the largest proportion of positive samples detected for hRSv (9.85% of all samples examined), a significant number of samples positive for hRv (2.76%), a smaller share found in isolated cases for hAdv, hBov, hCov, hMpv, hPiv types I and III (less than 1% of samples). At the same time, the content of hCov and hMpv in the 2021–2022 samples dropped to zero. Samples containing hPiv types II and IV were not detected during the period under the study. Co-infection between hRSv and hMpv viruses, as well as hAdv and hBov was founded only in individual cases (0.02% each).
The different hIv types were identified in 1,145 samples (24.30%). For over five years, hIv type B was found in only 14 samples (0.30%). Despite the fact, that the influenza virus was circulating in Kazakhstan in 2018 [15], no positive samples for influenza type B were found in our tested samples. Among 1,131 samples (24.00%) were positive for type A influenza, 575 samples (12.20%) belonged to A/H1N1 subtype, 312 samples (6.62%) were that of A/H3N2. We could not identify virus subtype in 244 samples (5.18%).
The genetic material of the detectable viruses was not found in 575 samples (12.20%).
Discussion
ARVIs, such as influenza and COVID-19, cause disorders in the respiratory tract and are of great importance for study. COVID-19 infection is associated with significant mortality, especially in high-risk groups (the elderly, people with chronic diseases, and healthcare providers). The gastrointestinal tract is believed to be the largest human immune organ and home to a complex community of commensal microorganisms. More recently, it has been assumed that people with comorbid chronic intestinal inflammation, when infected with this virus, tend to increase hypercytokinemia. This aggressive immune response leads to an increased risk of vascular hyperpermeability, multi-organ failure, and ultimately death. There is an opinion that chronic intestinal inflammation is associated with a specific composition of its microbiome, which is regulated by a diet. For example, a plant-based diet, such as vegetarianism, maintains a balanced gut microbiome. Thus, switching from an animal-based diet to a high-fiber diet can change the microbiota, which in turn will help suppress existing chronic inflammation and thereby reduce the development of a cytokine storm [26].
The highest incidence rates of the respiratory tract infections are recorded in childhood; this is due to the peculiarities of the immune response formation in children, the violation of microecological status in both the intestines and nasopharynx with a frequent formation of chronic inflammation foci [27,28,29]. The largest age group of working age patients (18–63 years old) is represented by 22.71% of samples; this is, primarily, because this age category frequently endures respiratory diseases of mild and moderate severity “out of bed”, seeking medical help only in cases of severe disease.
We did not examine the possibility of detecting multiple viruses in one patient simultaneously, because, in accordance with the quarantine measures the SARS-CoV-2-positive samples were prohibited from transportation outside medical laboratories in which they were detected. At the same time, there are data indicating a very low frequency of SARS-CoV-2 coinfection with other viral agents [30,31,32], which requires further study, since it was found that specific pre-existing immunity to seasonal coronaviruses can increase susceptibility to SARS-CoV-2 and predispose humans to a COVID-19 poor outcome [33]. According to data analysis of different influenza and SARS-CoV-2 co-infection, most cases of co-infection are associated with influenza A viruses, and the proportion of simultaneous infection with influenza viruses and SARS-CoV-2 in children was significantly higher than in adult patients, and the frequency of simultaneous infection was significantly higher in seriously ill individuals compared with other patients. In this connection, it is necessary to carefully monitor and study the concordance and interaction between SARS-CoV-2 and other respiratory viruses [34].
1145 samples (among 4,712 ones examined in RT-PCR) were positive for influenza, which is 24.30%, while a viral infection of non-influenza etiology was detected in 702 samples (14.90%). A high degree of the influenza spread was recorded by many researchers in different countries of the world [35,36,37,38]. At the same time, the co-circulation of different hIv variants including A/H1N1, A/H3N2, and type B [39] with a predominance of viruses with the antigenic formula A/H1N1was recorded both before the COVID-19 pandemic and during it [40].
When studying the etiological structure of ARVI of non-influenza etiology in Kazakhstan between 2016 and 2019, the predominance of hRSv (15.25%) and hRv (4.34%) was established, hPiv and hAdv were detected much less frequently (about 1%). hMpv, hBov, and hCov were revealed in isolated cases [14, 15]. The prevalence of hRSv and hRv in the pre-pandemic period was also recorded in different countries of Europe [41, 42], Asia [35, 37, 39, 43, 44], and Africa [36, 45].
Under the quarantine and social distancing measures in 2020–2022, despite the decrease in the total number of infected people, these viruses retained their leading positions among other viral agents. The hRSv proportion during the pandemic varied within a range of 2.41–3.14%, that of hRv within 0.87–1.69%, while the remaining sentinel viruses reached less than 1% and parainfluenza types II and IV was never identified. The highest coverage of the population with hRSv and hRv during the specified period was also recorded in other countries [46,47,48]. At the same time, several researchers have noticed the leading role of hMpv, hPiv as well as a significant contribution of hAdv in the general non-influenza viral pathology of the respiratory organs [35,36,37, 49, 50]. It is therefore possible to point out the territorial heterogeneity in the distribution of viral agents. In addition, there are the differences in the proportion of viruses depending on the age category of patients [35, 51].
The dynamics of the prevalence of pathogens causing non-influenza ARVIs and influenza during the period under the study and the proportion of viruses depending on the age category of patients are presented in Figs. 4, 5, and 6.
At the beginning of the period under consideration, before the COVID-19 pandemic, the total prevalence of detectable respiratory viruses increased from 46.51% in 2018 to 63.40% in 2019. There was further a sharp decline to 10.80% in 2020. In the subsequent 2021 and 2022, the level of occurrence of viruses that cause respiratory diseases among the population is gradually increasing (up to 21.98%) but did not reach the levels of 2018 and 2019.
The decline in the incidence of ARVIs and influenza in 2020 can be explained by the introduction of quarantine and self-isolation regimes during this period, a widespread rollout of anti-epidemic measures, enhanced measures to prevent ARVIs, and a decrease in the human migration inside and outside the country. In 2021–2022, a gradual easing of quarantine measures has begun and, as a result, a slow increase in the spread of pathogens was observed. Similar dynamics has also been observed in other countries that introduced social distancing and other anti-pandemic measures on their territories [46, 51].
Figure 7 shows statistics on the spread of SARS-CoV-2 in Kazakhstan from March 2020 to May 2022 [52]. A comparison of the prevalence rates of respiratory viruses and SARS-CoV-2 shows that against a sharp increase in SARS-CoV-2 in summer 2020 and tightening of the quarantine anti-pandemic regime, there was a sharp decline in the incidence of other ARVIs. With the beginning of the 2020–2021 epidemic period and easing of quarantine measures, there was a gradual increase in the incidence of both COVID-19 and other respiratory diseases of viral etiology, including influenza type A. It is estimated that the total incidence of ARVIs will gradually reach the pre-pandemic level, organically including, along with traditional respiratory viruses, the causative agent of COVID-19 [53].
Statistics on CoVID-19 spread in Kazakhstan during period from 13.03.2020 to 15.05.2022 [63]
In our studies of the 2021–2022 clinical samples, hCov was not detected although it was found that it was not displaced by the SARS-CoV-2 variant [54,55,56,57]; as previously, the simultaneous circulation of different variants of seasonal coronaviruses was observed [58,59,60,61]. It is possible that nonpharmaceutical interventions have influenced the prevalence of hCov and hMpv.
The effects of non-pharmaceutical interventions differ for each virus. For example, in South Korea, an increase in the spread of hBov was observed in late autumn and winter of 2020, when social distancing measures were eased [52]. Measures such as social distancing, wearing masks, etc., may have changed trends in seasonal respiratory virus outbreaks during the COVID-19 pandemic.
The world has been fighting the COVID-19 pandemic for more than two years by now. Due to its high susceptibility to mutations, a huge number of variants appeared, some of which had increased transmissibility, infectivity, lethality, and immune evasion. The greatest concern was caused by such virus variants as alpha, beta, gamma, and delta, which became more widespread. It would seem that the world has almost coped with the befallen humankind pandemic. However, the characteristics of the virus transmission and its high transmissibility with high mutagenicity raise concerns about the possibility of more dangerous variants to emerge in the future. Mass vaccination with modernized next-generation vaccines, taking into account circulating virus variants and observing safety measures, will help in the fight against the pandemic and the return of life to normal [62].
The analyze clinical samples collected from people who attended medical institutions in various regions of Kazakhstan to determine the etiology of viral pathogens and identify the dynamics of their spread was carried out in our studies. Unfortunately, for various reasons, we have not studied influenza coinfections/SARS-CoV-2 coinfection. Looking forward, we will continue analyzing disease surveillance data and its impact on transmission of various influenza lines and clinical trials of influenza coinfections/SARS-CoV-2 as well as keep monitoring changes in population immunity through serological tests This will contribute to new knowledge on the epidemiology of these and other pathogens and the development of new control capabilities.
Conclusion
In the current study, it was discovered that before the COVID-19 pandemic the total prevalence of detectable respiratory viruses was 46.51% in 2018 to 63.40 percent in 2019. In the bigining of COVID-19 pandemic, the rates of respiratory viruses sharply declined by 10.80%. Subsequently, in 2021–2022, the level of occurrence of respiratory diseases viruses among the population gradually increased. It was detected that most pathogens of respiratory infections of non-influenza etiology had respiratory syncytial virus, rhinovirus, adenovirus, paramyxovirus type I-III. Human coronaviruses, metapneumoviruses, bocaviruses were found less frequently. Whereas a circulation of different influenza virus variants (A/H1N1, A/H3N2 and type B) was detected both before and during the COVID-19 pandemic.
The above findings expand our knowledge of the significance of viruses in causing respiratory infections. Knowledge of the viral etiology of respiratory diseases can assist in recommending and implementing appropriate treatment strategies. Even though separate cases of ARVIs co-infection did not related with influenza and COVID-19 was found in our study, further research is necessary to determine whether multiple viral infections affect the disease severity.
References
Çelik I, Saatçi E, Eyüboğlu AF (2020) Emerging and reemerging respiratory viral infections up to Covid-19. Turk J Med Sci 50:557–562. https://doi.org/10.3906/sag-2004-126
Jain N, Lodha R, Kabra SK (2001) Upper respiratory tract infections. Indian J Pediatrics 68:1135–11358. https://doi.org/10.1007/BF02722930
Vikulov GKh (2019) Emerging and reemerging respiratory viral infections: algorithms of diagnostics, approaches of prophylaxis and therapy. Meditsina (Almaty) = Medicine (Almaty);7–8:53–64 (In Russian). https://doi.org/10.31082/1728-452X-2019-205-206-7-8-53-64
Influenza (Avian and other zoonotic). https://www.who.int/news-room/fact-sheets/detail/influenza-(avian-and-other-zoonotic; 2018 November 13. Accessed 12 July 2022
Karron RA, Black RE (2017) Determining the burden of respiratory syncytial virus disease: the known and the unknown. Lancet 390:917–918. https://doi.org/10.1016/s0140-6736(17)31476-9
Shchelkanov MYu, Popova AYu, Dedkov VG, et al. (2020) History of investigation and current classification of coronaviruses (Nidovirales: Coronaviridae). Russ J Infect Immun (Infektsiya i immunitet);10:221–246. https://doi.org/10.15789/2220-7619-HOI-1412
Nogales A, Aydillo T, Ávila-Pérez G et al (2019) Functional characterization and direct comparison of influenza A, B, C, and D NS1 proteins in vitro and in vivo. Front Microbiol 10:2862. https://doi.org/10.3389/fmicb.2019.02862
Payne S (2017) Family Orthomyxoviridae. In: Payne S (ed) Viruses: from understanding to investigation. Elsevier, Academic Press, Amsterdam, pp 197–208
Osterhaus AD, Rimmelzwaan GF, Martina BE et al (2000) Influenza B virus in seals. Science 288:1051–1053. https://doi.org/10.1126/science.288.5468.1051
Tsai CP, Tsai HJ (2019) Influenza B viruses in pigs Taiwan. Influenza Other Respir Viruses 13:91–105. https://doi.org/10.1111/irv.12588
Klivleyeva N, Ongarbayeva N, Korotetskiy I et al (2021) Coding-complete genome sequence of swine influenza virus isolate A/Swine/Karaganda/04/2020 (H1N1) from Kazakhstan. Microbiol Resource Announc. 10:e0078621. https://doi.org/10.1128/MRA.00786-21
Saktaganov NT, Klivleyeva NG, Ongarbayeva NS, et al. Study on antigenic relationships and biological properties of swine influenza A/H1N1 virus strains isolated in Northern Kazakhstan in 2018. Sel'skokhozyaistvennaya Biologiya [Agricultural Biology]. 2020;55:355–363. https://doi.org/10.15389/agrobiology.2020.2.355eng
Chiapponi C, Faccini S, De Mattia A et al (2016) Detection of influenza D virus among Swine and Cattle, Italy. Emerging Infect Dis 22:352–354. https://doi.org/10.3201/eid2202.151439
Klivleyeva NG, Lukmanova GV, Saktaganov NT et al. (2020) Acute respiratory viral infections in Kazakhstan in 2017–2019. Bull Natl Acad Sci Republic of Kazakhstan.; 3:29–35. https://doi.org/10.32014/2020.2518-1467.66 (ISSN 1991–3494
Klivleyeva NG, Ongarbayeva NS, Baimukhametova AM et al. (2021) Detection of influenza virus and pathogens of acute respiratory viral infections in population of Kazakhstan during 2018–2019 epidemic season. Russ J Infect Immun.;11:137–147. (In Russian) https://doi.org/10.15789/2220-7619-DOI-1348
Cebey-Lopez M, Herberg J, Pardo-Seco J et al (2015) Viral co-infections in pediatric patients hospitalized with lower tract acute respiratory infections. PLoS ONE 10:e0136526. https://doi.org/10.1371/journal.pone.0136526
Allander T, Tammi MT, Eriksson M et al (2005) Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci 102:12891–12896. https://doi.org/10.1073/pnas.0504666102
Taylor S, Lopez P, Weckx L et al (2017) Respiratory viruses and influenza-like illness: epidemiology and out comes in children aged 6 months to 10 years in a multi-country population sample. J Infect 74:29–41. https://doi.org/10.1016/j.jinf.2016.09.003
Ruohola A, Waris M, Allander T et al (2009) Viral etiology of common cold in children. Emerg Infect Dis 15:344–346. https://doi.org/10.3201/eid1502.081468
Kokoreva SP, Trushkina AV, Bolysheva GS, et al (2017) Etiological structure of acute respiratory viral infections in children in the epidemiological season 2014–2016. Infektsionnye bolezni = Infect Dis;15:133–134. (In Russian)
WHO. Manual for the laboratory diagnosis and virological surveillance of influenza. World Health Organization. https://apps.who.int/iris/bitstream/handle/10665/44518/9789241548090_eng.pdf?sequence=1&isAllowed=y. pdf; 2011 Accessed 18 September 2021
Zhang D, Mao H, Lou X et al (2018) Clinical evaluation of a panel of multiplex quantitative real-time reverse transcription polymerase chain reaction assays for the detection of 16 respiratory viruses associated with community-acquired pneumonia. Adv Virol 163:2855–2860. https://doi.org/10.1007/s00705-018-3921-8
Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR (2001) Universal primer set for the full-length amplification of all influenza A viruses. Adv Virol 146:2275–2289. https://doi.org/10.1007/s007050170002
Glantz S. Biomedical statistics. McGraw-Hill; 1994. (Translated to Russian. Moscow: Practice; 1998)
Statistical analysis in MS Excel. https://statanaliz.info/; Accessed 18 September 2021.
WHO. Pneumonia. https://www.who.int/ru/news-room/fact-sheets/detail/pneumonia; 2019 Accessed 02 September 2019
Rishi P, Thakur K, Vij S, Rishi L, Singh A, Kaur IP, Patel SKS, Lee JK, Kalia VC (2020) Diet, gut microbiota and COVID-19. Indian J Microbiol 60:420–429. https://doi.org/10.1007/s12088-020-00908-0
WHO (2018) Influenza. https://www.who.int/ru/news-room/fact-sheets/detail/influenza-(seasonal). Accessed 06 October 2018
Kanner EV, Maksimov ML, Ermolaeva AS et al (2018) Acute respiratory infections in children: features of the immune response and ways of correction. Russ Med J Med Rev 8:74–78 ((In Russian))
Marshall NC, Kariyawasam RM, Zelyas N et al (2021) Broad respiratory testing to identify SARS-CoV-2 viral co-circulation and inform diagnostic stewardship in the COVID-19 pandemic. Virol J 18:93. https://doi.org/10.1186/s12985-021-01545-9
Blyth CC, Webb SA, Kok J et al (2013) The impact of bacterial and viral co-infection in severe influenza. Influenza Other Respir Viruses 7:168–176. https://doi.org/10.1111/j.1750-2659.2012.00360.x
Basile K, Kok J, Dwyer DE (2018) Point-of-care diagnostics for respiratory viral infections. Expert Rev Mol Diagn 18:75–83. https://doi.org/10.1080/14737159.2018.1419065
Wratil PR, Schmacke NA, Karakoc B et al (2021) Evidence for increased SARS-CoV-2 susceptibility and COVID-19 severity related to pre-existing immunity to seasonal coronaviruses. Cell Rep 37:110169. https://doi.org/10.1016/j.celrep.2021.110169
Dao TL, Hoang VT, Colson P, Million M, Gautret P (2021) Co-infection of SARS-CoV-2 and influenza viruses: a systematic review and meta-analysis. J Clin Virol Plus. 1:100036. https://doi.org/10.1016/j.jcvp.2021.100036
Çiçek C, Arslan A, Karakuş HS, et al (2015) Akut solunum yolu enfeksiyonu olan hastalarda solunum viruslarının prevalansı ve mevsimsel dağılımı, 2002–2014. Mikrobiyoloji Bülteni.;49:188–200. (In Turkish). https://doi.org/10.5578/mb.9024
Ali A, Khowaja AR, Bashir MZ et al (2013) Role of human metapneumovirus, influenza a virus and respiratory syncytial virus in causing WHO-defined severe pneumonia in children in a developing country. PLoS ONE 8:e74756. https://doi.org/10.1371/journal.pone.0074756
Al-Romaihi HE, Smatti MK, Al-Khatib HA et al (2020) Molecular epidemiology of influenza, RSV, and other respiratory infections among children in Qatar: A six years report (2012–2017). Int J Infect Dis 95:133–141. https://doi.org/10.1016/j.ijid.2020.04.008
Liu T, Li Z, Zhang S et al (2015) Viral Etiology of acute respiratory tract infections in hospitalized children and adults in Shandong Province China. Virol J 12:168. https://doi.org/10.1186/s12985-015-0388-z
Naz R, Gul A, Javed U et al (2019) Etiology of acute viral respiratory infections common in Pakistan: a review. Rev Med Virol 29:2024. https://doi.org/10.1002/rmv.2024
Zhu G, Xu D, Zhang Y et al (2021) Epidemiological characteristics of four common respiratory viral infections in children. Virol J 18:10. https://doi.org/10.1186/s12985-020-01475-y
Garcia-Garcia ML, Calvo Rey C, Del Rosal RT (2016) Pediatric asthma and viral infection. Arch Bronconeumol 52:269–273. https://doi.org/10.1016/j.arbres.2015.11.008
Pisareva MM, Eder VA, Buzitskaya ZV, et al. (2018) Etiological structure of influenza and other ARVI in St. Petersburg during epidemic seasons 2012–2016. Voprosy Virusologii (Problems of Virology, Russian journal);;63:233–239. (In Russion). https://doi.org/10.18821/0507-4088-2018-63-5-233-239
Bashir UA, Nisar N, Mahmood N et al (2017) Molecular detection and characterization of respiratory syncytial virus B genotypes circulating in Pakistani children. Infect Genet Evol 47:125–131. https://doi.org/10.1016/j.meegid.2016.11.024
Khalid S, Ghani E, Ayyub M (2016) Study on etiology of viral lower respiratory tract infections in children under 10 years of age. J Virol Antiviral Res 5:4. https://doi.org/10.4172/2324-8955.1000160
Brini Khalifa I, Hannachi N, Guerrero A et al (2019) Demographic and seasonal characteristics of respiratory pathogens in neonates and infants aged 0 to 12 months in the Central-East region of Tunisia. J Med Virol 91:570–581. https://doi.org/10.1002/jmv.25347
Oh DY, Buda S, Biere B, et al. (2021) Trends in respiratory virus circulation following COVID-19-targeted nonpharmaceutical interventions in Germany, January–September 2020: Analysis of national surveillance data. The Lancet Regional Health – Europe.;6:100112. https://doi.org/10.1016/j.lanepe.2021.100112
Ljubin-Sternak S, Meštrović T, Lukšić I et al (2021) Seasonal coronaviruses and other neglected respiratory viruses: a global perspective and a local snapshot. Front Pub Health. 9:691163. https://doi.org/10.3389/fpubh.2021.691163
L'vov DK, Burtseva EI, Kolobukhina LV, et al. (2021) Peculiarities of the influenza and ARVI viruses circulation during epidemic season 2019–2020 in some regions of Russia. Voprosy Virusologii (Problems of Virology, Russian journal).;65:335–349. (In Russian). https://doi.org/10.36233/0507-4088-2020-65-6-4
Ji L, Chen L, Xu D et al (2021) Molecular typing and epidemiologic profiles of human metapneumovirus infection among children with severe acute respiratory infection in Huzhou, China. Mol Biol Rep 48:7697–7702. https://doi.org/10.1007/s11033-021-06776-1
Divarathna MVM, Rafeek RAM, Noordeen F (2020) A review on epidemiology and impact of human metapneumovirus infections in children using TIAB search strategy on PubMed and PubMed Central articles. Rev Med Virol 30:e2090. https://doi.org/10.1002/rmv.2090
Brestovac B, Lawrence C, Speers DJ et al (2020) Respiratory viral infections in Western Australians with cystic fibrosis. Respir Med 161:105854. https://doi.org/10.1016/j.rmed.2019.105854
Yum S, Hong K, Sohn S et al (2021) Trends in viral respiratory infections during COVID-19 Pandemic South Korea. Emerging Infectious Dis 27:1685–1688. https://doi.org/10.3201/eid2706.210135
Lagacé-Wiens P, Bullard J, Cole R, et al. (2021) Seasonality of coronaviruses and other respiratory viruses in Canada: implications for COVID-19. Canada Commun Dis Rep.;47:132–138. https://doi.org/10.14745/ccdr.v47i03a02
Biere B, Oh DY, Wolff T, et al. (2021) Surveillance of endemic human Coronaviruses in Germany, 2019/2020. The Lancet Regional Health – Europe.;11:100262. https://doi.org/10.1016/j.lanepe.2021.100262
Iannarella R, Lattanzi C, Cannata G, et al. (2020) Coronavirus infections in children: from SARS and MERS to COVID-19, a narrative review of epidemiological and clinical features. Acta Biomedica.;91:e2020032. https://doi.org/10.23750/abm.v91i3.10294
Yashavantha Rao HC, Jayabaskaran C (2020) The emergence of a novel coronavirus (SARS-CoV-2) disease and their neuroinvasive propensity may affect in COVID-19 patients. J Med Virol 92:786–790. https://doi.org/10.1002/jmv.25918
Zimmermann P, Curtis N (2020) coronavirus infections in children including COVID-19: an overview of the epidemiology, clinical features, diagnosis, treatment and prevention options in children. Pediatric Infect Dis J 39:355–368. https://doi.org/10.1097/INF.0000000000002660
Masse S, Capai L, Villechenaud N et al (2020) Epidemiology and clinical symptoms related to seasonal coronavirus identified in patients with acute respiratory infections consulting in primary care over six influenza seasons (2014–2020) in France. Viruses 12:630. https://doi.org/10.3390/v12060630
Corman VM, Lienau J, Witzenrath M (2019) Coronaviren als Ursache respiratorischer Infektionen [Coronaviruses as the cause of respiratory infections]. Internist (Berl) 60:1136–1145. https://doi.org/10.1007/s00108-019-00671-5
Kesheh MM, Hosseini P, Soltani S et al (2022) An overview on the seven pathogenic human coronaviruses. Rev Med Virol 32:e2282. https://doi.org/10.1002/rmv.2282
Park S, Lee Y, Michelow IC et al (2020) Global seasonality of human coronaviruses: a systematic review. Open Forum Infect Dis 7:ofaa44343. https://doi.org/10.1093/ofid/ofaa443
Thakur V, Bhola S, Thakur P, Patel SKS, Kulshrestha S, Ratho RK, Kumar P (2022) Waves and variants of SARS-CoV-2: understanding the causes and effect of the COVID-19 catastrophe. Infection 50:309–325. https://doi.org/10.1007/s15010-021-01734-2
Coronavirus. https://coronavirus-monitor.info/country/kazakhstan/; 2022 Accessed 08 July 2021.
Acknowledgements
We thank (G.G. Kasymova, A.K. Rachimbayeva, A.Zh. Murzagaliyeva, G.K. Xetayeva, R.T. Isabayeva, M.Y. Sagatova) as medical workers of the hospitals that helped in the collection of biological materials for this study.
Funding
This study was funded from a grant of the Program BR10965178 “Development of original domestic drugs with antiviral activity, effective against COVID-19 and influenza” provided by the Ministry of Education and Science of the Republic of Kazakhstan.
Author information
Authors and Affiliations
Contributions
Concept and design: NGK, GVL, TIG. Acquisition of data: NGK, GVL, TIG, MGS, NSO, NTS, AMB, SBB, and DAI. Analysis and interpretation of data: NGK, GVL, TIG. Drafting of the manuscript: NGK, GVL, and TIG. Critical revision of the manuscript for important intellectual content: all authors. Collection of biological materials: GGK, AKR, AZM, GKX, RTI, MYS.
Corresponding author
Ethics declarations
Conflicts of interest
All authors declare that they have no conflicts of interest.
Ethical Considerations
The study was carried out in the Laboratory for Biochemistry of Viruses at the LLP Research and Production Center for Microbiology and Virology (Almaty). The work with clinical samples was approved by the decision of the local ethics commission (Approval date 11.07.2022; Number: 03-01-01/128).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Klivleyeva, N., Lukmanova, G., Glebova, T. et al. Spread of Pathogens Causing Respiratory Viral Diseases Before and During CoVID-19 Pandemic in Kazakhstan. Indian J Microbiol 63, 129–138 (2023). https://doi.org/10.1007/s12088-023-01064-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-023-01064-x