Abstract
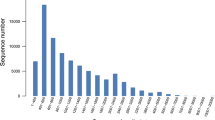
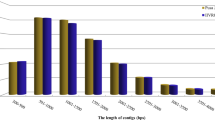
Stem gall (Protomyces macrosporus Unger), a serious disease that affects leaves, petioles, stems and fruits of coriander (Coriandrum sativum L.) causing heavy loss in yield. Genetic improvement of coriander for stem gall disease is indispensable. Coriander cultivars of stem gall resistance (ACr-1) and susceptible (CS-6) leaf samples were utilized and transcriptome sequenced using Illumina NextSeq500 platform. After trimming low-quality reads and adapter sequences, a total of 49,163,108 and 43,746,120 high-quality reads were retained and further assembly resulted validated transcripts of 59,933 and 56,861. We have predicted 52,506 and 48,858 coding sequences (CDS) of which 50,506 and 46,945 were annotated using NCBI nr database. Gene ontology analysis annotated 19,099 and 17,625 terms; pathway analysis obtained 24 different functional pathway categories; signal transduction, transport, catabolism, translation and carbohydrate metabolism pathways etc. were dominated. Differentially expressed genes analysis predicted 13,123 CDS commonly expressed of which 431 and 400 genes were significantly upregulated and downregulated, respectively, in which R genes, stress inducible transcription factors such as ERF, NAC, bZIP, MYB, DREB and WRKY and antifungal related genes were predicted. The real-time PCR analysis of HSP20 gene expression in resistance showed upregulation by 10-fold over susceptible sample and 18s used as a housekeeping gene for normalization. The present results provide an insights into various aspects underlying the development of resistance to stem gall in coriander.





Similar content being viewed by others
References
Altschul S. F., Gish W., Miller W., Myers E. W. and Lipman D. J. 1990 Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Anders S. and Huber W. 2010 Differential expression analysis for sequence count data. Genome Biol. 11, R106.
Bell-Lelong D. A., Cusumano J. C., Meyer K. and Chapple C. 1997 Cinnamate-4-hydroxylase expression in Arabidopsis. Regulation in response to development and the environment. Plant Physiol. 113, 729–738.
Bolger A. M., Lohse M. and Usadel B. 2014 Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120.
Conesa A. and Gotz S. 2008 Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics. https://doi.org/10.1155/2008/619832.
Choudhary S., Sharma R. and Verma A. K. 2017 Microsatellite in coriander: a cross species amplification within Apiaceae family. Int. J. Curr. Microbiol. App. Sci. 5, 2714–2721.
Das K., Prasanna R. and Saxena A. K. 2017 Rhizobia: a potential biocontrol agent for soilborne fungal pathogens. Folia Microbiol. (Praha) 62, 425–435.
Dangl J. L. and McDowell J. M. 2006 Two modes of pathogen recognition by plants. Proc. Natl. Acad. Sci. USA 103, 8575–8576.
De Lucca A. J. 2000 Antifungal peptides: potential candidates for the treatment of fungal infections. Expert Opin. Investig. Drugs 9, 273-299.
DeYoung B. J. and Innes R. W. 2006 Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7, 1243–1249.
Diederichsen A. 1996 Coriander (Coriandrum sativum L.). Promoting the conservation and use of underutilized and neglected crops. 3. Institute of Plant Genetics and Crop Plant Research, Gatersleben/International Plant Genetic Resources Institute, Rome.
Dixon R. A. and Paiva N. L. 1995 Stress-induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097.
Galata M., Sarker L. S. and Mahmoud S. S. 2014 Transcriptome profiling, and cloning and characterization of the main monoterpene synthases of Coriandrum sativum L. Phytochemistry 102, 64–73.
Gutterson N. and Reuber T. L. 2004 Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7, 465–471.
Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J. et al. 2013 De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512.
Hamel L. P., Nicole M. C., Duplessis S. and Ellis B. E. 2012 Mitogen-activated protein kinase signaling in plant-interacting fungi: distinct messages from conserved messengers. Plant Cell 24, 1327–1351.
Heidrich K., Wirthmueller L., Tasset C., Pouzet C., Deslandes L. and Parker J. E. 2011 Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334, 1401–1404.
Howe E. A., Sinha R., Schlauch D. and Quackenbush J. 2011 RNA-Seq analysis in MeV. Bioinformatics 27, 3209–3210.
Huckelhoven R. 2007 Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45, 101–127.
Kanehisa M. and Goto S. 2000 KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30.
Li H. and Durbin R. 2009 Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760.
Lopes-Caitar V. S., de Carvalho M. C., Darben L. M., Kuwahara M. K., Nepomuceno A. L., Dias W. P. et al. 2013 Genome-wide analysis of the Hsp20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genomics 14, 577.
Malinovsky F. G., Fangel J. U. and Willats W. G. 2014 The role of the cell wall in plant immunity. Front. Plant Sci. 5, 178.
Malhotra S. K., Kakani R. K., Sharma Y. K., Saxena S. N. and Vashishtha B. B. 2016a Comparative superiority of coriander variety NRCSS ACr-1 for yield and stem gall disease tolerance. Indian J. Hortic. 73, 453–455.
Malhotra S. K., Kakani R. K., Sharma Y. K. and Singh D. K. 2016b Ajmer coriander-1 (NRCSS, Acr-1), resistant to stem gall disease-an innovative farming technology. Indian J. Arecanut Spices Med. Pl. 18, 1–7.
Marrs K. A. 1996 The functions and regulation of glutathione S-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 127–158.
McGrath K. C., Dombrecht B., Manners J. M., Schenk P. M., Edgar C. I, Maclean D. J. et al. 2005 Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139, 949–959.
Moriya Y., Itoh M., Okuda S., Yoshizawa A. C. and Kanehisa M. 2007 KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185.
Naika M., Shameer K. and Sowdhamini R. 2013a Comparative analyses of stress-responsive genes in Arabidopsis thaliana: insight from genomic data mining, functional enrichment, pathway analysis and phenomics. Mol. Biosyst. 9, 1888–1908.
Naika M., Shameer K., Mathew O. K., Gowda R. and Sowdhamini R. 2013b STIFDB2 an updated version of plant stress-responsive transcription factor database with additional stress signals, stress-responsive transcription factor binding sites and stress-responsive genes in Arabidopsis and rice. Plant Cell Physiol. 54, e8.
Nicot N., Hausman J. F., Hoffmann L. and Evers D. 2005 Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 56, 2907–2914.
Nene Y. L. and Kharbanda P. D. 1967 Studies on the control of stem gall of corianders International Symposium of Plant Pathology (Dec. 27–Jan. 1967), pp. 61 (Abstr.). Indian Phytopathological Society, New Delhi.
Rutter G. A., Da Silva Xavier G. and Leclerc I. 2003 Roles of 5’-AMP-activated protein kinase (AMPK) in mammalian glucose homoeostasis. Biochem. J. 375, 1–16.
Sahib N. G., Anwar F., Gilani A. H., Hamid A. A., Saari N. and Alkharfy K. M. 2013 Coriander (Coriandrum sativum L.): a potential source of high-value components for functional foods and nutraceuticals-a review. Phytother. Res. 27, 1439–1456.
Schulz M. H., Zerbino D. R., Vingron M. and Birney E. 2012 Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28, 1086–1092.
Shrivastava H. C. 1955 Studies on tumour disease of Coriandrum sativum caused by Protomyces macrosporus Unger, pp. 218. Proceedings of the 42nd Indian Science Congress part III. Baroda.
Strickler S. R., Bombarely A. and Mueller L. A. 2012 Designing a transcriptome next-generation sequencing project for a nonmodel plant species. Am. J. Bot. 99, 257–266.
Sydow H. and Butler E. J. 1911 Fungi India Orientalis. Ann. Mycol. 9, 372–421.
Takahashi A., Casais C., Ichimura K. and Shirasu K. 2003 HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 100, 11777–11782.
Thomma B. P., Eggermont K., Tierens K. F. and Broekaert W. F. 1999 Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121, 1093–1102.
To J. P., Deruere J., Maxwell B. B., Morris V. F., Hutchison C. E., Ferreira F. J. et al. 2007 Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell 19, 3901–3914.
Vossen J. H., Abd-El-Haliem A., Fradin E. F., van den Berg G. C., Ekengren S. K., Meijer H. J. et al. 2010 Identification of tomato phosphatidylinositol-specific phospholipase-C (PI-PLC) family members and the role of PLC4 and PLC6 in HR and disease resistance. Plant J. 62, 224–239.
Wang Z., Gerstein M. and Snyder M. 2009 RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63.
Yang Y., Shah J. and Klessig D. F. 1997 Signal perception and transduction in plant defense responses. Genes Dev. 11, 1621–1639.
Yang K., Liu Y. and Zhang S. 2001 Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. 98, 741–746.
Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z. et al. 2006 WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34, W293–W297.
Zerbino D. R. 2010 Using the Velvet de novo assembler for short-read sequencing technologies. Curr. Protoc. Bioinformatics 11, 11–15.
Zhao Q. Y., Wang Y., Kong Y. M., Luo D., Li X. and Hao P. 2011 Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics 12.
Zheng Z., Qamar S. A., Chen Z. and Mengiste T. 2006 Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605.
Acknowledgements
We thank Dr Gopal Lal, Director, NRCSS for timely support and encouragement during the research work. The research is supported by the ICAR-NRC on Seed Spices, Ajmer, Rajasthan, India. SC, RS, RDM, GL thank ICAR-NRC on Seed Spices, Ajmer for providing all the facilities. MBNN thank UHS, Bagalkot for infrastructure facilities.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Corresponding editor: H. A. Ranganath
SC conceived theme of the study. RS, RDM and RS Performed the experiments and collected samples. MBNN and SC analysed the data and drafted the manuscript. GL, RS, RDM and RS edited the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Choudhary, S., Naika, M.B.N., Sharma, R. et al. Transcriptome profiling of coriander: a dual purpose crop unravels stem gall resistance genes. J Genet 98, 19 (2019). https://doi.org/10.1007/s12041-019-1064-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-019-1064-7