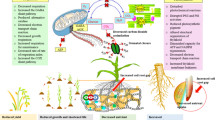
Abstract
Different stresses include nutrient deficiency, pathogen attack, exposure to toxic chemicals etc. Transcriptomic studies have been mainly applied to only a few plant species including the model plant, Arabidopsis thaliana. These studies have provided valuable insights into the genetic networks of plant stress responses. Transcriptomics applied to cash crops including barley, rice, sugarcane, wheat and maize have further helped in understanding physiological and molecular responses in terms of genome sequence, gene regulation, gene differentiation, posttranscriptional modifications and gene splicing. On the other hand, comparative transcriptomics has provided more information about plant’s response to diverse stresses. Thus, transcriptomics, together with other biotechnological approaches helps in development of stress tolerance in crops against the climate change.
Similar content being viewed by others
References
Amiour N., Imbaud S., Clement G., Agier N., Zivy M., Valot B. et al. 2012 The use of metabolomics integrated with transcriptomic and proteomic studies for identifying key steps involved in the control of nitrogen metabolism in crops such as maize . J. Exp. Bot. 63, 5017–5033.
An Y. and Lin L. 2011 Transcriptional regulatory programs underlying barley germination and regulatory functions of gibberellin and abscisic acid. BMC Plant Biol. 11, 105.
Armengaud P., Breitling R. and Amtmann A. 2004 The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol. 136, 2556–2576.
Baerson S. R., Sanchez-Moreiras A., Pedrol-Bonjoch N., Schulz M., Kagan I. A., Agarwal A. K. et al. 2005 Detoxification and transcriptome response in Arabidopsis seedlings exposed to the allelochemical benzoxazolin-2(3H)-one. J. Biol. Chem. 280, 21867–21881.
Bai X., Rivera-Vega L., Mamidala P., Bonello P., Herms D. A. and Mittapalli O. 2011 Transcriptomic signatures of Ash (Fraxinus spp.) phloem. PLoS One 6, e16368.
Barrero J. M., Talbot M. J., White R. G., Jacobsen J. V. and Gubler F. 2009 Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley. Plant Physiol. 150, 1006–1021.
Barros E., Lezar S., Anttonen M. J., Van Dijk J. P., Rohlig R. M., Kok E. J. et al. 2010 Comparison of two GM maize varieties with a near-isogenic non-GM variety using transcriptomics, proteomics and metabolomics. Plant Biotechnol. J. 8, 436–451.
Barros N. M., Mauro S. M. Z., di Laia M. L., de Dabbas K. M., Ferro J. A. and Ferro M. I. T. 2004 Differential expression pattern in sugar cane varieties infected with Leifsonia xyli subsp xyli. The international conference on the status of plant and animal genome research, 12, Jan.10–14. San Diego, USA.
Bock K. W., Honys D., Ward J. M., Padmanaban S., Nawrocki E. P., Hirschi K. D. et al. 2006 Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol. 140, 1151–1168.
Borges F., Gomes G., Gardner R., Moreno N., McCormick S., Feijo J. A. et al. 2008 Comparative transcriptomics of Arabidopsis sperm Cells. Plant Physiol. 148, 1168–1181.
Brady S. M., Long T. A. and Benfey P. N. 2006 Unraveling the dynamic transcriptome. Plant Cell 18, 2101–2111.
Brautigam A., Kajala K., Wullenweber J., Sommer M., Gagneul D., Weber K. L. et al. 2011 An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 155, 142–156.
Brenner S., Johnson M., Bridgham J., Golda G., Lloyd D. H., Johnson D. et al. 2000 Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18, 630–634.
Casati P., Campi M., Morrow D. J., Fernandes J. F. and Walbot V. 2011a Transcriptomic, proteomic and metabolomic analysis of UV-B signaling in maize. BMC Genomics 12, 321.
Casati P., Morrow D. J., Fernandes J. and Walbot V. 2011b UV-B signaling in maize: transcriptomic and metabolomic studies at different irradiation times. Plant Signal Behav. 6, 1926–1931.
Casu R. E., Hotta C. T. and Souza G. M. 2010 Functional genomics: transcriptomics of sugarcane–current status and future prospects. In Genetics genomics and breeding of sugarcane (ed. R. J. Henry and C. Kole), pp. 167–191. Science Publishers, Enfield, USA.
Chang Y. M., Liu W. Y., Shih A. C., Shen M. N., Lu C. H., Lu M. Y. et al. 2012 Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol. 160, 165–177.
Chaudhary B., Hovav R., Rapp R., Verma N., Udall J. A. and Wendel J. F. 2008 Global analysis of gene expression in cotton fibers from wild and domesticated Gossypium barbadense. Evol. Dev. 10, 567–582.
Chelaifa H., Monnier A. and Ainouche M. 2010 Transcriptomic changes following recent natural hybridization and allopolyploidy in the salt marsh species Spartina × townsendii and Spartina anglica (Poaceae). New Phytol. 186, 161–174.
Cho K., Agrawal G. K., Shibato J., Jung Y. H., Kim Y. K., Nahm B. H. et al. 2007 Survey of differentially expressed proteins and genes in jasmonic acid treated rice seedling shoot and root at the proteomics and transcriptomics levels. J. Proteomics Res. 6, 3581–3603.
Cho K., Shibato J., Agrawal G. K., Jung Y. H., Kubo A., Jwa N. S. et al. 2008 Integrated transcriptomics, proteomics, and metabolomics analyses to survey ozone responses in the leaves of rice seedling. J. Proteom. Res. 7, 2980–2998.
Cho K., Shibato J., Kubo A., Kohno Y., Satoh K., Kikuchi S. et al. 2013 Genome-wide mapping of the ozone-responsive transcriptomes in rice panicle and seed tissues reveals novel insight into their regulatory events. Biotechnol. Lett. 35, 647–656.
Clark T. A., Sugnet C. W. and Ares Jr M. 2002 Genome wide analysis of mRNA processing in yeast using splicing specific microarrays. Science 296, 907–910.
Cloonan N., Forrest A. R., Kolle G., Gardiner B. B., Faulkner G. J., Brown M. K. et al. 2008 Stem cell transcriptome profiling via massive scale mRNA sequencing. Nat. Methods 5, 613–619.
Cohen D., Bogeat-Triboulot M. B., Tisserant E., Balzergue S., Martin-Magniette M. L., Lelandais G. et al. 2010 Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genome 11, 630.
Contento A. L., Kim S. J. and Bassham D. C. 2004 Transcriptome profiling of the response of Arabidopsis suspension culture cells to suc starvation. Plant Physiol. 135, 2330–2347.
Coram T. E., Settles M. L. and Chen X. 2009 Large-scale analysis of antisense transcription in wheat using the Affymetrix Gene Chip Wheat Genome Array. BMC Genome 10, 253.
Crismani W., Baumann U., Sutton T., Shirley N., Webster T., Spangenberg G. et al. 2006 Microarray expression analysis of meiosis and microsporogenesis in hexaploid bread wheat. BMC Genome 7, 267.
Crismani W., Kapoor S. and Able J. A. 2011 Comparative transcriptomics reveals 129 transcripts that are temporally regulated during anther development and meiotic progression in both bread wheat (Triticum aestivum) and rice (Oryza sativa). Int. J. Plant Genome (article ID 931898).
Dassanayake M., Haas J. S., Bohnert H. J. and Cheeseman J. M. 2009 Shedding light on an extremophile lifestyle through transcriptomics. New Phytol. 183, 764–775.
David L., Huber W., Granovskaia M., Toedling J., Palm C. J., Bofkin L. et al. 2006 A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. USA 103, 5320– 5325.
De Vos M., Van Oosten V. R., Van Poecke R. M., Van Pelt J. A., Pozo M. J., Mueller M. J. et al. 2005 Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 18, 923–937.
Desikan R., Mackerness S. A. H., Hancock J. T. and Neill S. J. 2001 Regulation of Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127, 159–172.
Diaz P., Betti M., Sanchez D. H., Udvardi M. K., Monza J. and Marquez A. J. 2010 Deficiency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytol. 188, 1001–1013.
Dos Santos P. B., Soares-Cavalcanti N. M., Vieira-de-Melo G. S. and Benko-Iseppon A. M. 2011 Osmoprotectants in the sugarcane (Saccharum spp.) transcriptome revealed by in silico evaluation. Comput. Intel. Meth. Bioinfo. Biostat. 6685, 44–58.
Ekman D. R., Lorenz W. W., Przybyla A. E., Wolfe N. L. and Dean J. F. D. 2003 SAGE analysis of transcriptome responses in Arabidopsis roots exposed to 2,4,6-trinitrotoluene. Plant Physiol. 133, 1397–1406.
Eulgem T. 2004 Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10, 71–78.
Evers D., Legay S., Lamoreux D., Hausman J. F., Hoffmann L. and Renaut J. 2012 Towards a synthetic view of potato cold and salt stress response by transcriptomic and proteomic analyses. Plant Mol. Biol. 78, 503–514.
Felix J. M., Papini-Terzi F. S., Rocha F. R., Vencio R. Z. N., Vicentini R., Nishiyama Jr M. Y. et al. 2009 Expression profile of signal transduction components in a sugarcane population segregating for sugar content. Tropic Plant Biol. 2, 98–109.
Fowler S. and Thomashow M. F. 2002 Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690.
Gao F., Jordan M. C. and Ayele B. T. 2012 Transcriptional programs regulating seed dormancy and its release by after-ripening in common wheat (Triticum aestivum L.) Plant Biotechnol. J. 10, 465–476.
Gao S., Zhang H., Tian Y., Li F., Zhang Z., Lu X. et al. 2008 Expression of TERF1 in rice regulates expression of stress-responsive genes and enhances tolerance to drought and high-salinity. Plant Cell Rep. 27, 1787–1795.
Gardiner S. A., Boddu J., Berthiller F., Hametner C., Stupar R. M., Adam G. et al. 2010 Transcriptome analysis of the barley–deoxynivalenol interaction: evidence for a role of glutathione in deoxynivalenol detoxification. Mol. Plant Microbe Interact. 23, 962–976.
Ge X., Chen W., Song S., Wang W., Hu S. and Yu J. 2008 Transcriptomic profiling of mature embryo from an elite super-hybrid rice LYP9 and its parental lines. BMC Plant Biol. 8, 114.
Gerhard D. S., Wagner L., Feingold E. A., Shenmen C. M., Grouse L. H., Schuler G. et al. 2004 The status, quality, and expansion of NIH full length cDNA project: the mammalian gene collection. Genome Res. 14, 2121–2127.
Gomase V. S. and Tagore S. 2008 Transcriptomics. Curr. Drug Metabol. 9, 245–249.
Gulick P. J., Drouin S., Yu Z., Danyluk J., Poisson G., Monroy A. F. et al. 2005 Transcriptome comparison of winter and spring wheat responding to low temperature. Genome 48, 913–923.
Hayes K. R., Beatty M., Meng X., Simmons C. R., Habben J. E. and Danilevskaya O. N. 2010 Maize global transcriptomics reveals pervasive leaf diurnal rhythms but rhythms in developing ears are largely limited to the core oscillator. PLoS One 5, e12887.
Hermans C., Vuylsteke M., Coppens F., Craciun A., Inze D. and Verbruggen N. 2010 Early transcriptomic changes induced by magnesium deficiency in Arabidopsis thaliana reveal the alteration of circadian clock gene expression in roots and the triggering of abscisic acid-responsive genes. New Phytol. 187, 119–131.
Hocher V., Alloisio N., Auguy F., Fournier P., Doumas P., Pujic P. et al. 2011 Transcriptomics of actinorhizal symbioses reveals homologs of the whole common symbiotic signaling cascade. Plant Physiol. 156, 700–711.
Holt R. A. and Jones S. J. 2008 The new paradigm of flow cell sequencing. Genome Res. 18, 839–846.
Honys D. and Twell D. 2003 Comparative analysis of the arabidopsis pollen transcriptome. Plant Physiol. 132, 640–652.
Honys D. and Twell D. 2004 Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5, R85.
Hovav R., Udall J. A., Hovav E., Rapp R., Flagel L. and Wendel J. F. 2008 A majority of cotton genes are expressed in single-celled fiber. Planta 227, 319–329.
Huang G., Xu W., Gong S., Li B., Wang X., Xu D. et al. 2008 Characterization of 19 novel cotton FLA genes and their expression profiling in fiber development and in response to phytohormones and salt stress. Physiol. Plant. 134, 348–359.
Ibarra-Laclette E., Albert V. A., Perez-Torres C. A., Zamudio-Hernandez F., Ortega-Estrada M. J., Herrera-Estrella A. et al. 2011 Transcriptomics and molecular evolutionary rate analysis of the bladderwort (Utricularia), a carnivorous plant with a minimal genome. BMC Plant Biol. 11, 101.
Ilut D. C., Coate J. E., Luciano A. K., Owens T. G., May G. D., Farmer A. et al. 2012 A comparative transcriptomic study of an allotetraploid and its diploid progenitors illustrates the unique advantages and challenges of RNA-seq in plant species. Am. J. Bot. 99, 383–396.
Jiang S. Y., González J. M. and Ramachandran S. 2013 Comparative genomic and transcriptomic analysis of tandemly and segmentally duplicated genes in rice. PLoS One 8, e63551.
Jukanti A. K., Heidlebaugh N. M., Parrott D. L., Fischer I. A., Mclnnerney K. and Fischer A. M. 2008 Comparative transcriptome profiling of near-isogenic barley (Hordeum vulgare) lines differing in the allelic state of a major grain protein content locus identifies genes with possible roles in leaf senescence and nitrogen reallocation. New Phytol. 177, 333–349.
Kahlau S. and Bock R. 2008 Plastid transcriptomics and translatomics of tomato fruit development and chloroplast to chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Physiol. 20, 856–874.
Kempema L. A., Cui X., Holzer F. M. and Walling L. L. 2007 Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in response to aphids. Plant Physiol. 143, 849–865.
Kodzius R., Kojima M., Nishiyori H., Nakamura M., Fukuda S., Taqami M. et al. 2006 CAGE: cap analysis of gene expression. Nat. Methods 3, 211–222.
Koenig D., Jiménez-Gómez J. M., Kimura S., Fulop D., Chitwood D. H., Headland L. R. et al. 2013 Comparative transcriptomics reveal patterns of selection in domesticated and wild tomato. Proc. Natl. Acad. Sci. USA 110, E2655–E2662.
Kreps J. A., Wu Y., Chang H. S., Zhu T., Wang X. and Harper J. F. 2002 Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130, 2129–2141.
Lee B., Henderson D. A. and Zhu J. K. 2005 The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17, 3155–3175.
Lee S., Lee K., Kim K., Choi G. J., Yoon S. H., Ji H. C. et al. 2009 Identification of salt-stress induced differentially expressed genes in barley leaves using the annealing control-primer-based GeneFishing technique. Afr. J. Biotechnol. 8, 1326–1331.
Lembke C. G., Nishiyama Jr. M. Y., Sato P. M., de Andrade R. F. and Souza G. M. 2012 Identification of sense and antisense transcripts regulated by drought in sugarcane. Plant Mol. Biol. 79, 461–477.
Li L., Qiu X., Li X., Wang S., Zhang Q. and Lian X. 2010 Transcriptomic analysis of rice responses to low phosphorus stress. Chinese Sci. Bull. 55, 251–258.
Li T., Li H., Zhang Y. and Liu J. 2010 Identification and analysis of seven H2 O 2-responsive miRNAs and 32 new miRNAs in the seedlings of rice (Oryza sativa L. ssp. indica). Nucleic Acid Res. 39, 2821–2833.
Li Y., Sun C., Huang C., Pan J., Wang L. and Fan X. 2009 Mechanisms of progressive water deficit tolerance and growth recovery of chinese maize foundation genotypes Huangzao 4 and Chang 7-2, which are proposed on the basis of comparison of physiological and transcriptomic responses. Plant Cell Physiol. 50, 2092–2111.
Liu C., Li S., Wang M. and Xia G. 2012 A transcriptomic analysis reveals the nature of salinity tolerance of a wheat introgression line. Plant Mol. Biol. 78, 159–169.
Liu M. S., Li H. C., Lai Y. M., Lo H. F. and Chen L. F. 2013 Proteomics and transcriptomics of broccoli subjected to exogenously supplied and transgenic senescence-induced cytokinin for amelioration of postharvest yellowing. J. Proteom. 93, 133–144.
Maathius F. J. M. 2006 The role of monovalent cation transporters in plant responses to salinity. J. Exp. Bot. 57, 1137–1147.
Manners J. M. and Casu R. E. 2011 Transcriptome analysis and functional genomics of sugarcane. Tropic Plant Biol. 4, 9–21.
Maruyama-Nakashita A., Inoue E., Watanabe-Takahashi A., Yamaya T. and Takahashi H. 2003 Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol. 132, 597–605.
Mattiello L., Kirst M., Da Silva F. R., Jorge R. A. and Menossi M. 2010 Transcriptional profile of maize roots under acid soil growth. BMC Plant Biol. 10, 196.
Mortazavi A., Williams B. A., McCue K., Schaeffer L. and Wold B. 2008 Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628.
Muller R., Morant M., Jarmer H., Nilsson L. and Neilson T. H. 2007 Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 143, 156–171.
Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M. et al. 2008 The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349.
Narsai R., Howell K. A., Carroll A., Ivanova A., Millar A. H. and Whelan J. 2009 Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol. 151, 306–322.
Nikiforova V., Freitag J., Kempa S., Adamik M., Hesse H. and Hoefgen R. 2003 Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J. 33, 633–650.
Nodine M. D. and Bartel D. P. 2012 Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482, 94–97.
Norton G. J., Lou-Hing D. E., Meharg A. A. and Price A. H. 2008 Rice–arsenate interactions in hydrophonics: whole genome transcriptional analysis. J. Exp. Bot. 59, 2267–2276.
Nowrousian M. 2007 Of patterns and pathways: microarray technologies for the analysis of filamentous fungi. Fungal Biol. Rev. 21, 171–178.
Okoniewski M. J. and Miller C. J. 2006 Hybridization interactions between probesets in short oligo microarrays lead to spurious correlations. BMC Bioinformatics 7, 276.
Omrane S., Ferrarini A., D’Apuzzo E., Rogato A., Delledonne M. and Chiurazzi M. 2009 Symbiotic competence in Lotus japonicus is affected by plant nitrogen status: transcriptomic identification of genes affected by a new signalling pathway. New Phytol. 183, 380–394.
Padmalatha K. V., Dhandapani G., Kanakachari M., Kumar S., Dass A., Patil D. P. et al. 2012 Genome-wide transcriptomic analysis of cotton under drought stress reveal significant down-regulation of genes and pathways involved in fibre elongation and up-regulation of defense responsive genes. Plant Mol. Biol. 78, 223–246.
Pallavicini A., Del Terra L., Sondhal M. R., Guerreiro-Filho O., Asquini E., Martellosi C. et al. 2005 Transcriptomics of resistance response in Coffea arabica L. In Proceedings of the ASIC 2004, pp. 584–590. 20th International Conference on Coffee Science, Bangalore, India.
Papini-Terzi F. S., Rocha F. R., Vêncio R. Z., Oliveira K. C., Felix Jde M., Vicentini R. et al. 2005 Transcription profiling of signal transduction related genes in sugarcane tissues. DNA Res. 12, 27–38.
Park W., Scheffler B. E., Bauer P. J. and Campbell B. T. 2012 Genome-wide identification of differentially expressed genes under water deficit stress in upland cotton (Gossypium hirsutum L.). BMC Plant Biol. 12, 90.
Parrott D. L., Mclnnerney K., Feller U. and Fischer A. M. 2007 Steam-girdling of barley (Hordeum vulgare) leaves leads to carbohydrate accumulation and accelerated leaf senescence, facilitating transcriptomic analysis of senescence-associated genes. New Phytol. 176, 56–69.
Patade V. Y., Bhargava S. and Suprasanna P. 2012 Transcript expression profiling of stress responsive genes in response to short-term salt or PEG stress in sugarcane leaves. Mol. Biol. Rep. 39, 3311–3318.
Pellny T. K., Lovegrove A., Freeman J., Tosi P., Love C. G., Knox P. et al. 2012 Cell walls of developing wheat starchy endosperm: comparison of composition and RNA-seq transcriptome. Plant Physiol. 158, 612–627.
Pina C., Pinto F., Feijo J. A. and Becker J. D. 2005 Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 138, 744–756.
Pufky J., Qiu Y., Rao M. V., Hurban P. and Jones A. M. 2003 The auxin-induced transcriptome for etiolated Arabidopsis seedlings using a structure/function approach. Funct. Integr. Genomics 3, 135–143.
Queval G., Neukermans J., Vanderauwera S., Van Breusegem F. and Noctor G. 2012 Day length is a key regulator of transcriptomic responses to both CO2 and H2O2 in Arabidopsis. Plant Cell Environ. 35, 374–387.
Rapp R. A., Haigler C. H., Flagel L., Hovav R. H., Udall J. A. and Wendel J. F. 2010 Gene expression in developing fibres of upland cotton (Gossypium hirsutum L.) was massively altered by domestication. BMC Biol. 8, 139.
Rodrigues F. A., De Laia M. L. and Zingaretti S. M. 2009 Analysis of gene expression profiles under water stress in tolerant and sensitive sugarcane plants. Plant Sci. 176, 286–302.
Royce T. E., Rozowsky J. S. and Gerstein M. B. 2007 Toward a universal microarray: prediction of gene expression through nearest neighbor probe sequence identification. Nucleic Acid Res. 35, e99.
Ruzicka D. R., Hausmann N. T., Barrios-Masias F. H., Jackson L. E. and Schachtman D. P. 2012 Transcriptomic and metabolic responses of mycorrhizal roots to nitrogen patches under field conditions. Plant Soil 350, 145–162.
Sanchez-Pons N., Irar S., Garcia-Muniz N. and Vicient C. M. 2011 Transcriptomic and proteomic profiling of maize embryos exposed to camptothecin. BMC Plant Biol. 11, 91.
Schenk P. M., Carvalhais L. C. and Kazan K. 2012 Unraveling plant–microbe interactions: can multi-species transcriptomics help? Trend Biotechnol. 30, 177–184.
Shanker A. K., Maheswari M., Yadav S. K., Desai S., Bhanu D., Attal N. B. et al. 2014 Drought stress responses in crops. Funct. Integr. Genom. 14, 11–22.
Slocombe S. P., Schauvinhold I., McQuinn R. P., Besser K., Welsby N. A., Harper A. et al. 2008 Transcriptomic and reverse genetic analyses of branched chain fatty acid and acyl sugar production in Solanum pennellii and Nicotiana benthamiana. Plant Physiol. 148, 1830–1846.
Spieb N., Oufir M., Matusikova I., Stierschneider M., Kopecky D., Homolka A. et al. 2012 Ecophysiological and transcriptomic responses of oak (Quercus robur) to long-term drought exposure and rewatering. Environ. Exp. Bot. 77, 117–126.
Strickler S. R., Bombarely A. and Mueller L. A. 2012 Designing a transcriptome next-generation sequencing project for a nonmodel plant species. Am. J. Bot. 99, 257–266.
Suzuki H., Rodriguez-Uribe L., Xu J. and Zhang J. 2013 Transcriptome analysis of cytoplasmic male sterility and restoration in CMS-D8 cotton. Plant Cell Rep. 32, 1531–1542.
Szakasits D., Heinen P., Weiczorek K., Hofmann J., Wagner F., Kreil D. P. et al. 2009 The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant J. 57, 771–784.
Tan K. C., Ipcho S. V. S., Trengove R. D., Oliver R. P. and Solomon P. S. 2009 Assessing the impact of transcriptomics, proteomics and metabolomics on fungal phytopathology. Mol. Plant Pathol. 10, 703–715.
Thompson G. A. and Goggin F. L. 2006 Transcriptomics and functional genomics of plant defence induction by phloem feeding insects. J. Exp. Biol. 57, 755–766.
Vera J. C., Wheat C. W., Fescemyer H. W., Frilander M. J., Crawford D. L., Hanski I. et al. 2008 Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol. Encol. 17, 1636–1647.
Vieira-de-Melo G. S., dos Santos P. B., Soares-Cavalcanti N. M. and Benko-Iseppon A. M. 2011 Identification and expression of early nodulin in sugarcane transcriptome revealed by in silico analysis. Comput. Intel. Meth. Bioinfo. Biostat. 6685, 72–85.
Walter S. and Doohan F. 2011 Transcript profiling of the phytotoxic response of wheat to the Fusarium mycotoxin deoxynivalenol. Mycotoxin Res. 27, 221–230.
Wan Y., Poole R. L., Huttly A. K., Toscano-Underwood C., Feeney K., Welham S. et al. 2008 Transcriptome analysis of grain development in hexaploid wheat. BMC Genomics 9, 121.
Wang H., Wan A., Hsu C., Lee K., Yu S. and Jauh G. 2007 Transcriptomic adaptations in rice suspension cells under sucrose starvation. Plant Mol. Biol. 63, 441–463.
Wang X., Elling A. A., Li X., Li N., Peng Z., He G. et al. 2009 Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21, 1053–1069.
Wang Y., Deng D., Ding H., Xu X., Zhang R., Wang S. et al. 2013 Gibberellin biosynthetic deficiency is responsible for maize dominant dwarf11 (d11) mutant phenotype: physiological and transcriptomic evidence. PLoS One 8, e66466.
Wang Y., Zhang W., Song L., Zou J., Su Z. and Wu W. 2008 Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148, 1201–1211.
Wang Z., Gerstein M. and Snyder M. 2009 RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63.
Wasaki J., Shinano T., Onishi K., Yonetani R., Yazaki J., Fujii F. et al. 2006 Transcriptomic analysis indicates putative metabolic changes caused by manipulation of phosphorus availability in rice leaves. J. Exp. Bot. 57, 2049–2059.
Wasaki J., Yonetani R., Kuroda S., Shinano T., Yazaki J., Fujii F. et al. 2003 Transcriptomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant Cell Environ. 26, 1515–1523.
Wei M., Song M., Fan S. and Yu S. 2013 Transcriptomic analysis of differentially expressed genes during anther development in genetic male sterile and wild type cotton by digital gene-expression profiling. BMC Genomics 14, 97.
Wilson I. D., Barker G. L. A., Lu C., Coghill J. A., Beswick R. W., Lenton J. R. et al. 2005 Alteration of the embryo transcriptome of hexaploid winter wheat (Triticum aestivum cv. Mercia) during maturation and germination. Func. Integr. Genomics. 5, 144–154.
Wu J. 2010 Transcriptomic analysis of barley plant responses to cold stress. Dissertation submitted in partial fulfillment of the Master of Research (MRes) in advanced genomic and proteomic sciences, School of Biosciences, The University of Nottingham.
Xin Z., Zhao Y. and Zheng Z. 2005 Transcriptome analysis reveals specific modulation of abscisic acid signaling by ROP10 small GTPase in Arabidopsis. Plant Physiol. 139, 1350–1365.
Xu X. H., Chen H., Sang Y. L., Wang F., Ma J. P., Gao X. et al. 2012 Identification of genes specifically or preferentially expressed in maize silk reveals similarity and diversity in transcript abundance of different dry stigmas. BMC Genomics 13, 294.
Xu Z., Zhang C., Zhang X., Liu C., Wu X., Yang Z. et al. 2013 Transcriptome profiling reveals auxin and cytokinin regulating somatic embryogenesis in different sister lines of cotton cultivar CCRI24. J. Integr. Plant Biol. 55, 631–642.
Yamada K., Lim J., Dale J. M., Chen H., Shinn P., Palm C. J. et al. 2003 Emperical analysis of transcriptional activity in the Arabidopsis genome. Science 302, 842–846.
Yano R., Takebayashi Y., Nambara E., Kamiya Y. and Seo M. 2013 Combining association mapping and transcriptomics identify HD2B histone deacetylase as a genetic factor associated with seed dormancy in Arabidopsis thaliana. Plant J. 74, 815–828.
Yu H., Hogan P. and Sundaresan V. 2005 Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol. 139, 1853–1869.
Zeimann M., Kamboj A., Hove R. M., Loveridge S., El-Osta A. and Bhave M. 2013 Analysis of the barley leaf transcriptome under salinity stress using mRNA-Seq. Acta Physiol. Plant. 35, 1915–1924.
Zou X., Jiang Y., Liu L., Zhang Z. and Zheng Y. 2010 Identification of transcriptome induced in roots of maize seedlings at the late stage of waterlogging. BMC Plant Biol. 10, 189.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Imadi S. R., Kazi A. G., Ahanger M. A., Gucel S. and Ahmad P. 2015 Plant transcriptomics and responses to environmental stress: an overview. J. Genet. 94, xx–xx]
Rights and permissions
About this article
Cite this article
IMADI, S.R., KAZI, A.G., AHANGER, M.A. et al. Plant transcriptomics and responses to environmental stress: an overview. J Genet 94, 525–537 (2015). https://doi.org/10.1007/s12041-015-0545-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-015-0545-6