Abstract
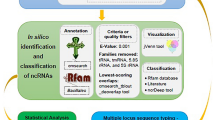
Proteus vulgaris is a rod-shaped Gram-negative bacterium known to be the member of Enterobacteriaceae that is able to cause disease in human being. Generally, non-protein-coding RNAs (npcRNAs) do not code for proteins, but they play a vital role in gene regulation at the RNA level including pathogenicity. The present study aims at elucidating homologous npcRNAs from other bacteria in Proteus vulgaris. A comparative genomic analysis was carried out to identify npcRNA homolog of other Enterobacteriaceae pathogens in Proteus vulgaris. A total of 231 npcRNAs previously reported in Salmonella typhi, Salmonella typhimurium and Escherichia coli were screened using BLASTn tool against Proteus vulgaris genome. Interestingly, 33 npcRNAs are homologs to Proteus vulgaris. Northern blot analysis of 6 out of 33 npcRNA candidates confirmed their expression and showed that most of them are differentially expressed during lag, exponential and stationary growth phases. This study is the first approach of identification and characterization of npcRNAs in P. vulgaris. Hence, this could be a pioneer study to further validate the regulatory functions of these npcRNAs to fill the gaps in understanding of the pathogenicity of P. vulgaris.



Similar content being viewed by others
References
Antoni R 1997 Potential virulence factors of Proteus Bacilli. Microbiol. Mol. Biol. Rev. 61 65–89
Argaman L, Hershberg R, Vogel J, et al. 2001 Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11 941–950
Broomfield RJ, Morgan SD, Khan A and Stickler DJ 2009 Crystalline bacterial biofilm formation on urinary catheters by urease-producing urinary tract pathogens: a simple method of control. J. Med. Microbiol. 58 1367–1375
Carver T, Harris SR, Berriman M, Parkhill J and McQuillan JA 2011 Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28 464–469
Chinni SV, Raabe CA, Zakaria R, et al. 2010 Experimental identification and characterization of 97 novel npcRNA candidates in Salmonella enterica serovar Typhi. Nucleic Acids Res. 38 5893–5908
Coornaert A, Chiaruttini C, Springer M and Guillier M 2013 Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLoS Genet. 9 e1003156
Gopalan V, Vioque A and Altman S 2002 RNase P: variations and uses. J. Biol. Chem. 277 6759–6762
Göpel Y, Papenfort K, Reichenbach B, Vogel J and Görke B 2013 Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Genes Dev. 27 552–564
Hartmann E and Hartmann RK 2003 The enigma of ribonuclease P evolution. Trends Genet. 19 561–569
Herbig A and Nieselt K 2011 nocoRNAc: characterization of non-coding RNAs in prokaryotes. BMC Bioinform. 12 40
Hiasa H and Marians KJ 1992 Differential inhibition of the DNA translocation and DNA unwinding activities of DNA helicases by the Escherichia coli Tus protein. J. Biol. Chem. 267 11379–11385
Hill TM and Marians KJ 1990 Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc. Natl. Acad. Sci. USA 87 2481–2485
Hill TM, Tecklenburg ML, Pelletier AJ and Kuempel PL 1989 tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc. Natl. Acad. Sci. USA 86 1593–1597
Huang L, Hu J, Su Y, Qin Y, et al. 2016 Genome-wide detection of predicted non-coding RNAs related to the adhesion process in Vibrio alginolyticus using high-throughput sequencing. Front. Microbiol.
Hughes WH 1957 A reconsideration of the swarming of Proteus vulgaris. Microbiology 17 49–58
Jørgensen MG, Nielsen JS, Boysen A, Franch T, Møller-Jensen J and Valentin-Hansen P 2012 Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol. Microbiol. 84 36–50
Kalamorz F, Reichenbach B, März W, Rak B and Görke B 2007 Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol. Microbiol. 65 1518–1533
Kawano M, Reynolds AA, Miranda-Rios J and Storz G 2005 Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 33 1040–1050
Kery MB, Feldman M, Livny J and Tjaden B 2014 TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res. 42 W124–W129
Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M and John B 2010 A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 38 e98–e98
KishanRaj S, Sumitha S, Siventhiran B, et al. 2018 In silico ‘fishing’ using known small regulatory RNA (sRNA) candidates as the decoy from Escherichia coli, Salmonella typhi and Salmonella typhimurium manifested 14 novel sRNA candidates in the orthologous region of Proteus mirabilis. Mol. Biol. Rep.
Li R, Zhu H and Luo Y 2016 Understanding the functions of long non-coding RNAs through their higher-order structures. Int. J Mol. Sci. 17 702
Li Y and Altman S 2003 A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc. Natl. Acad. Sci. 100 13213–13218
Matsunaga J and Haake DA 2018 cis-acting determinant limiting expression of sphingomyelinase gene sph2 in Leptospira interrogans, identified with a gfp reporter plasmid. Appl Env. Microbiol. 84 e02068-18
Minogue TD, Daligault HE, Davenport KW, et al. 2014 Draft genome assemblies of Proteus mirabilis ATCC 7002 and Proteus vulgaris ATCC 49132. Genome Announc. 2 e01064-e1114
Mordi RM and Momoh MI 2008 Incidence of Proteus species in wound infections and their sensitivity pattern in the University of Benin Teaching Hospital. Afr. J Biotechnol. 8 725–730
Musa H, Kasim FH, Gunny AAN, Gopinath SC, Chinni SV and Ahmad MA 2019 Whole genome sequence of moderate halophilic marine bacterium Marinobacter litoralis SW-45: abundance of non-coding RNAs. Int. J. Biol. Macromol. 133 1288–1298
Muto A, Sato M, Tadaki T, Fukushima M, Ushida C and Himeno H 1996 Structure and function of 10Sa RNA: trans-translation system. Biochimie 78 985–991
Nithya R, Ahmed SA, Hoe C-H, et al. 2015 Non-protein coding RNA genes as the novel diagnostic markers for the discrimination of Salmonella species using PCR. PLoS One 10 e0118668
Papenfort K and Vogel J 2009 Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res. Microbiol. 160 278–287
Pelletier AJ, Hill TM and Kuempel PL 1989 Termination sites T1 and T2 from the Escherichia coli chromosome inhibit DNA replication in ColE1-derived plasmids. J. Bacteriol. 171 1739–1741
Per Johnsson 2013 Evolutionary conservation of long non-coding RNAs; sequence, structure function (Elsevier) pp. 1063–1071
Pulvermacher SC, Stauffer LT and Stauffer GV 2009 Role of the sRNA GcvB in regulation of cycA in Escherichia coli. Microbiology 155 106–114
Raabe CA, Sanchez CP, Randau G, et al. 2009 A global view of the nonprotein-coding transcriptome in Plasmodium falciparum. Nucleic Acids Res. 38 608–617
Ranquet C and Gottesman S 2007 Translational regulation of the Escherichia coli stress factor RpoS: a role for SsrA and Lon. J. Bacteriol. 189 4872–4879
Urbanowski ML, Stauffer LT and Stauffer GV 2000 The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 37 856–868
Vijian D, Chinni SV, Yin LS, Lertanantawong B and Surareungchai W 2016 Non-protein coding RNA-based genosensor with quantum dots as electrochemical labels for attomolar detection of multiple pathogens. Biosens. Bioelectron. 77 805–811
Vogel J and Sharma CM 2005 How to find small non-coding RNAs in bacteria. Biol. Chem. 386 1219–1238
Wassarman KM and Storz G 2000 6S RNA regulates E coli RNA polymerase activity. Cell 101 613–623
Yean CYS, Raju KS, Xavier R, Subramaniam S, Gopinath SC and Chinni SV 2016 Molecular detection of methicillin-resistant Staphylococcus aureus by non-protein coding RNA-mediated monoplex polymerase chain reaction. PLoS ONE 11 e0158736
Acknowledgements
This work was supported by FRGS, Malaysia, FRGS/1/2018/STG03/AIMST/02/1.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
KishanRaj, S., Sumitha, S., Tang, TH. et al. Comparative genomic identification and characterization of npcRNA homologs in Proteus vulgaris. J Biosci 46, 108 (2021). https://doi.org/10.1007/s12038-021-00230-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12038-021-00230-x