Abstract
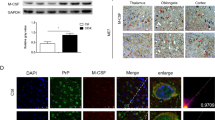
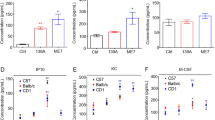
Interleukin 3 (IL-3) plays an important role in hematopoiesis and immune regulation, brain IL-3/IL-3R signaling has been shown to involve in the physiological and pathological processes of a variety of neurodegenerative diseases, but its role in prion diseases is rarely described. Here, the changes of IL-3/IL-3R and its downstream signaling pathways in a scrapie-infected cell line and in the brains of several scrapie-infected rodent models were evaluated by various methods. Markedly decreased IL-3Rα were observed in the brains of scrapie-infected rodents at terminal stage and in the prion-infected cell model, which showed increased in the brain samples collected at early and middle stage of infection. The IL-3 levels were almost unchanged in the brains of scrapie-infected mice and in the prion-infected cell line. Morphological assays identified close co-localization of the increased IL-3Rα signals with NeuN- and Iba1-positive cells, whereas co-localization of IL-3 signals with NeuN- and GFAP-positive cells in the scrapie-infected brain tissues. Some downstream components of IL-3/IL-3R pathways, including JAK2-STAT5 and PI3K/AKT/mTOR pathways, were downregulated in the brains of scrapie-infected rodents at terminal stage and in the prion-infected cells. Stimulation of recombinant IL-3 on the cultured cells showed prion that the prion-infected cells displayed markedly more reluctant responses of JAK2-STAT5 and PI3K/AKT/mTOR pathways than the normal partner cells. These data suggest that although prion infection or PrPSc accumulation in brain tissues does not affect IL-3 expression, it significantly downregulates IL-3R levels, thereby inhibiting the downstream pathways of IL-3/IL-3R and blocking the neuroregulatory and neuroprotective activities of IL-3.















Similar content being viewed by others
Data Availability
The datasets generated for this study are available on request to the corresponding.
References
Baldwin KJ, Correll CM (2019) Prion disease. Semin Neurol 39(4):428–439. https://doi.org/10.1055/s-0039-1687841
Prusiner SB (1998) Prions. Proc Natl Acad Sci U S A 95(23):13363–13383. https://doi.org/10.1073/pnas.95.23.13363
Wadsworth JD, Collinge J (2011) Molecular pathology of human prion disease. Acta Neuropathol 121(1):69–77. https://doi.org/10.1007/s00401-010-0735-5
Aguzzi A, Zhu C (2017) Microglia in prion diseases. J Clin Invest 127(9):3230–3239. https://doi.org/10.1172/JCI90605
Mindur JE, Swirski FK (2019) Growth factors as immunotherapeutic targets in cardiovascular disease. Arterioscler Thromb Vasc Biol 39(7):1275–1287. https://doi.org/10.1161/ATVBAHA.119.311994
Appel K, Buttini M, Sauter A, Gebicke-Haerter PJ (1995) Cloning of rat interleukin-3 receptor beta-subunit from cultured microglia and its mRNA expression in vivo. J Neurosci 15(8):5800–5809. https://doi.org/10.1523/JNEUROSCI.15-08-05800.1995
Natarajan C, Sriram S, Muthian G, Bright JJ (2004) Signaling through JAK2-STAT5 pathway is essential for IL-3-induced activation of microglia. Glia 45(2):188–196. https://doi.org/10.1002/glia.10316
Kiddle SJ, Thambisetty M, Simmons A, Riddoch-Contreras J, Hye A, Westman E, Pike I, Ward M et al (2012) Plasma based markers of [11C] PiB-PET brain amyloid burden. PLoS ONE 7(9):e44260. https://doi.org/10.1371/journal.pone.0044260
Zambrano A, Otth C, Maccioni RB, Concha II (2010) IL-3 controls tau modifications and protects cortical neurons from neurodegeneration. Curr Alzheimer Res 7(7):615–624. https://doi.org/10.2174/156720510793499011
Frendl G, Beller DI (1990) Regulation of macrophage activation by IL-3. I. IL-3 functions as a macrophage-activating factor with unique properties, inducing Ia and lymphocyte function-associated antigen-1 but not cytotoxicity. J Immunol 144(9):3392–3399
Frendl G (1992) Interleukin 3: from colony-stimulating factor to pluripotent immunoregulatory cytokine. Int J Immunopharmacol 14(3):421–430. https://doi.org/10.1016/0192-0561(92)90172-h
Frei K, Bodmer S, Schwerdel C, Fontana A (1986) Astrocyte-derived interleukin 3 as a growth factor for microglia cells and peritoneal macrophages. J Immunol 137(11):3521–3527
McAlpine CS, Park J, Griciuc A, Kim E, Choi SH, Iwamoto Y, Kiss MG, Christie KA et al (2021) Astrocytic interleukin-3 programs microglia and limits Alzheimer’s disease. Nature 595(7869):701–706. https://doi.org/10.1038/s41586-021-03734-6
Clarke MC, Haig DA (1971) Multiplication of scrapie agent in mouse spleen. Res Vet Sci 12(2):195–197
Birkett CR, Hennion RM, Bembridge DA, Clarke MC, Chree A, Bruce ME, Bostock CJ (2001) Scrapie strains maintain biological phenotypes on propagation in a cell line in culture. EMBO J 20(13):3351–3358. https://doi.org/10.1093/emboj/20.13.3351
Shi Q, Zhang BY, Gao C, Zhang J, Jiang HY, Chen C, Han J, Dong XP (2012) Mouse-adapted scrapie strains 139A and ME7 overcome species barrier to induce experimental scrapie in hamsters and changed their pathogenic features. Virol J 9:63. https://doi.org/10.1186/1743-422X-9-63
Hu C, Chen C, Xia Y, Chen J, Yang W, Wang L, Chen D-D, Wu Y-Z et al (2022) Different Aberrant changes of mglur5 and its downstream signaling pathways in the scrapie-infected cell line and the brains of scrapie-infected experimental rodents. Front Cell Dev Biol 10. https://doi.org/10.3389/fcell.2022.844378
Huang SCZ, Yu JF, Young D, Bashey A, Ho AD, Law P (1999) Correlation between IL-3 receptor expression and growth potential of human CD34+ hematopoietic cells from different tissues. Stem Cells 17(5):265–272. https://doi.org/10.1002/stem.170265
Fogha J, Bayry J, Diharce J, de Brevern AG (2021) Structural and evolutionary exploration of the IL-3 family and its alpha subunit receptors. Amino Acids 53(8):1211–1227. https://doi.org/10.1007/s00726-021-03026-3
Zambrano A, Otth C, Mujica L, Concha II, Maccioni RB (2007) Interleukin-3 prevents neuronal death induced by amyloid peptide. BMC Neurosci 8:82. https://doi.org/10.1186/1471-2202-8-82
Wen TC, Tanaka J, Peng H, Desaki J, Matsuda S, Maeda N, Fujita H, Sato K et al (1998) Interleukin 3 prevents delayed neuronal death in the hippocampal CA1 field. J Exp Med 188(4):635–649. https://doi.org/10.1084/jem.188.4.635
Frei K, Bodmer S, Schwerdel C, Fontana A (1985) Astrocytes of the brain synthesize interleukin 3-like factors. J Immunol 135(6):4044–4047
Konishi Y, Kamegai M, Takahashi K, Kunishita T, Tabira T (1994) Production of interleukin-3 by murine central nervous system neurons. Neurosci Lett 182(2):271–274. https://doi.org/10.1016/0304-3940(94)90814-1
Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR et al (1997) Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science 275(5300):661–665. https://doi.org/10.1126/science.275.5300.661
Chen C, Shi Q, Xiao K, Zhou W, Gao C, Gao L, Han J, Wang J et al (2022) Activation of innate immunity and autophagy in brain tissues with prion disease and degradation of abnormal PrPs in Cells - China’s Studies. China CDC Wkly 4(33):735–740. https://doi.org/10.46234/ccdcw2022.153
Shi Q, Chen C, Xiao K, Zhou W, Gao C, Gao L, Han J, Wang J et al (2022) Extensive disturbances of intracellular components and dysfunctions of biological pathways in the brain tissues during prion infection - China’s Studies. China CDC Wkly 4(33):741–747. https://doi.org/10.46234/ccdcw2022.154
Chen J, Chen C, Hu C, Liu L, Xia Y, Wang L, Yang W, Wu HY et al (2020) IP10, KC and M-CSF are remarkably increased in the brains from the various strains of experimental mice infected with different scrapie agents. Virol Sin 35(5):614–625. https://doi.org/10.1007/s12250-020-00216-3
Xia Y, Chen C, Chen J, Hu C, Yang W, Wang L, Liu L, Gao L-P et al (2022) Enhanced M-CSF/CSF1R signaling closely associates with PrPSc Accumulation in the scrapie-infected cell line and the brains of scrapie-infected experimental rodents. Mol Neurobiol 59(10):6534–6551. https://doi.org/10.1007/s12035-022-02989-y
Abedpoor N, Taghian F, Hajibabaie F (2022) Cross brain-gut analysis highlighted hub genes and LncRNA networks differentially modified during leucine consumption and endurance exercise in mice with depression-like behaviors. Mol Neurobiol 59(7):4106–4123. https://doi.org/10.1007/s12035-022-02835-1
Funding
This work was supported by SKLID Development Grant (2021SKLID504, 2019SKLID401, 2019SKLID603).
Author information
Authors and Affiliations
Contributions
XXJ and CC contributed to study design, performed assays and data analysis, and prepared the manuscript. CH, ZYC, and WWZ assisted with the assays of cultured cells. WYZ, QF, RHA, and XL assisted with WB and IFA analysis assays. KX and QS assisted with statistical analysis. CC and XPD contributed to design, study concept, and manuscript preparation.
Corresponding authors
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jia, XX., Chen, C., Hu, C. et al. Abnormal Changes of IL3/IL3R and Its Downstream Signaling Pathways in the Prion-Infected Cell Line and in the Brains of Scrapie-Infected Rodents. Mol Neurobiol (2023). https://doi.org/10.1007/s12035-023-03511-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12035-023-03511-8