Abstract
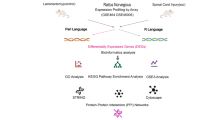
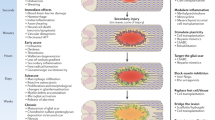
The pathology of spinal cord injury (SCI), including primary and secondary injuries, primarily involves hemorrhage, ischemia, edema, and inflammatory responses. Cell transplantation has been the most promising treatment for SCI in recent years; however, its specific molecular mechanism remains unclear. In this study, bioinformatics analysis verified by experiment was used to elucidate the hub genes associated with SCI and to discover the underlying molecular mechanisms of cell intervention. GSE46988 data were downloaded from the Gene Expression Omnibus dataset. In our study, differentially expressed genes (DEGs) were reanalyzed using the “R” software (R v4.2.1). Functional enrichment and protein–protein interaction network analyses were performed, and key modules and hub genes were identified. Network construction was performed for the hub genes and their associated miRNAs. Finally, a semi-quantitative analysis of hub genes and pathways was performed using quantitative real-time polymerase chain reaction. In total, 718 DEGs were identified, mainly enriched in immune and inflammation-related functions. We found that Cd4, Tp53, Rac2, and Akt3 differed between vehicle and transplanted groups, suggesting that these genes may play an essential role in the transplantation of olfactory ensheathing cells, while a toll-like receptor signaling pathway was significantly enriched in Gene set enrichment analysis, and then, the differences were statistically significant by experimentally verifying the expression of their associated molecules (Tlr4, Nf-κb, Ikkβ, Cxcl2, and Tnf-α). In addition, we searched for upstream regulatory molecules of these four central genes and constructed a regulatory network. This study is the first to construct a regulatory network for olfactory ensheathing cell transplantation in treating SCI, providing a new idea for SCI cell therapy.









Similar content being viewed by others
Data Availability
The data used to support the findings of this study have been included in this article.
Abbreviations
- SCI:
-
Spinal cord injury
- GSE:
-
GEO series
- DEGs:
-
Differentially expressed genes
- PPI:
-
Protein–protein interaction network
- GSEA:
-
Gene set enrichment analysis
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- Tp53:
-
Tumor suppressor p53
- Rac2:
-
Rac family small GTPase 2
- OECs:
-
Olfactory ensheathing cells
- miRNA:
-
MicroRNA
- mRNA:
-
Messenger RNA
- KEGG:
-
Kyoto Encyclopedia of Genes and Genome
- HE:
-
Hematoxylin and eosin
- SEM:
-
Mean standard error
References
Eckert MJ, Marti MJ (2017) Trauma: spinal cord injury. Surg Clin North Am 97(5):1031–1045
Ibrahim E, Brackett NL, Lynne CM (2016) Advances in the management of infertility in men with spinal cord injury. Asian J Androl 18(3):382–90
Witiw CD, Fehlings MG (2015) Acute spinal cord injury. J Spinal Disord Tech 28(6):202–210
Lee BB et al (2014) The global map for traumatic spinal cord injury epidemiology: update 2011, global incidence rate. Spinal Cord 52(2):110–116
Yang R et al (2017) Epidemiological characteristics of traumatic spinal cord injury in Guangdong, China. Spine (Phila Pa 1976) 42(9):E555-e561
Chen J et al (2021) Epidemiological features of traumatic spinal cord injury in Guangdong Province, China. J Spinal Cord Med 44(2):276–281
Assinck P et al (2017) Cell transplantation therapy for spinal cord injury. Nat Neurosci 20(5):637–647
Radtke C, Kocsis JD (2014) Olfactory-ensheathing cell transplantation for peripheral nerve repair: update on recent developments. Cells Tissues Organs 200(1):48–58
Gu J et al (2019) Olfactory ensheathing cells promote nerve regeneration and functional recovery after facial nerve defects. Neural Regen Res 14(1):124–131
Tsai S, Gamblin TC (2019) Gamblin, molecular characteristics of biliary tract and primary liver tumors. Surg Oncol Clin N Am 28(4):685–693
Yan P et al (2018) In silico analyses for potential key genes associated with gastric cancer. Peer J 6:e6092
Ye B et al (2018) High-throughput sequencing of the immune repertoire in oncology: applications for clinical diagnosis, monitoring, and immunotherapies. Cancer Lett 416:42–56
Çakmak HA, Demir M (2020) MicroRNA and cardiovascular diseases. Balkan Med J 37(2):60–71
Chen L et al (2018) Proteomics for biomarker identification and clinical application in kidney disease. Adv Clin Chem 85:91–113
Su W et al (2021) Exploring the pathogenesis of psoriasis complicated with atherosclerosis via microarray data analysis. Front Immunol 12:667690
Li R, Sim I (2019) How clinical trial data sharing platforms can advance the study of biomarkers. J Law Med Ethics 47(3):369–373
Subramanian A et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102(43):15545–15550
Debrabant B (2017) The null hypothesis of GSEA, and a novel statistical model for competitive gene set analysis. Bioinformatics 33(9):1271–1277
Szklarczyk D et al (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45(D1):D362-d368
Franceschini A et al (2013) STRING v91: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41(Database issue):D808-15
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4:2
de Rivero Vaccari JP et al (2008) A molecular platform in neurons regulates inflammation after spinal cord injury. J Neurosci 28(13):3404–3414
Hasegawa T et al (2023) Cytotoxic CD4(+) T cells eliminate senescent cells by targeting cytomegalovirus antigen. Cell 186(7):1417-1431.e20
Itano AA, Jenkins MK (2003) Antigen presentation to naive CD4 T cells in the lymph node. Nat Immunol 4(8):733–739
Kane LP, Lin J, Weiss A (2000) Signal transduction by the TCR for antigen. Curr Opin Immunol 12(3):242–249
Fu Y et al (2023) Identification and validation of immune-related genes diagnostic for progression of atherosclerosis and diabetes. J Inflamm Res 16:505–521
Gay D et al (1987) Functional interaction between human T-cell protein CD4 and the major histocompatibility complex HLA-DR antigen. Nature 328(6131):626–629
Klatzmann DR, McDougal JS, Maddon PJ (1990) The CD4 molecule and HIV infection. Immunodefic Rev 2(1):43–66
Gebhardt T et al (2011) Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature 477(7363):216–219
Laumaea A et al (2023) Small CD4 mimetics sensitize HIV-1-infected macrophages to antibody-dependent cellular cytotoxicity. Cell Rep 42(1):111983
Diallo MS et al (2021) A comparison of cell activation, exhaustion, and expression of HIV coreceptors and restriction factors in HIV-1- and HIV-2-infected nonprogressors. AIDS Res Hum Retroviruses 37(3):214–223
Yan H et al (2022) Design of a bispecific HIV entry inhibitor targeting the cell receptor CD4 and viral fusion protein Gp41. Front Cell Infect Microbiol 12:916487
Donehower LA et al (2019) Integrated analysis of TP53 gene and pathway alterations in the cancer genome atlas. Cell Rep 28(5):1370-1384.e5
Sablina AA et al (2005) The antioxidant function of the p53 tumor suppressor. Nat Med 11(12):1306–1313
Kotipatruni RR et al (2011) p53- and Bax-mediated apoptosis in injured rat spinal cord. Neurochem Res 36(11):2063–2074
Mehta SL et al (2013) Overexpression of human selenoprotein H in neuronal cells enhances mitochondrial biogenesis and function through activation of protein kinase A, protein kinase B, and cyclic adenosine monophosphate response element-binding protein pathway. Int J Biochem Cell Biol 45(3):604–611
Kanno H et al (2012) The role of mTOR signaling pathway in spinal cord injury. Cell Cycle 11(17):3175–3179
Wennerberg K, Der CJ (2004) Rho-family GTPases: it’s not only Rac and Rho (and I like it). J Cell Sci 117(Pt 8):1301–1312
Didsbury J et al (1989) rac, a novel ras-related family of proteins that are botulinum toxin substrates. J Biol Chem 264(28):16378–16382
Hall A, Lallin G (2010) Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb Perspect Biol 2(2):a001818
Yang Z et al (2017) Identification of crucial genes associated with rat traumatic spinal cord injury. Mol Med Rep 15(4):1997–2006
Iversen PO et al (2000) Depressed immunity and impaired proliferation of hematopoietic progenitor cells in patients with complete spinal cord injury. Blood 96(6):2081–2083
Dooley JL et al (2009) Regulation of inflammation by Rac2 in immune complex-mediated acute lung injury. Am J Physiol Lung Cell Mol Physiol 297(6):L1091–L1102
Tian Y, Autieri MV (2007) Cytokine expression and AIF-1-mediated activation of Rac2 in vascular smooth muscle cells: a role for Rac2 in VSMC activation. Am J Physiol Cell Physiol 292(2):C841–C849
Spillane M, Gallo G (2014) Involvement of Rho-family GTPases in axon branching. Small GTPases 5:e27974
Numano F et al (2009) Critical involvement of Rho GTPase activity in the efficient transplantation of neural stem cells into the injured spinal cord. Mol Brain 2:37
Lundquist EA (2003) Rac proteins and the control of axon development. Curr Opin Neurobiol 13(3):384–390
Carmona FJ et al (2016) AKT signaling in ERBB2-amplified breast cancer. Pharmacol Ther 158:63–70
Sarbassov DD et al (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307(5712):1098–1101
Alessi DR et al (1997) Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol 7(4):261–269
Walker KS et al (1998) Activation of protein kinase B beta and gamma isoforms by insulin in vivo and by 3-phosphoinositide-dependent protein kinase-1 in vitro: comparison with protein kinase B alpha. Biochem J 331(Pt 1):299–308
Meier R et al (1997) Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase Bbeta. J Biol Chem 272(48):30491–30497
Recabarren D, Alarcón M (2017) Gene networks in neurodegenerative disorders. Life Sci 183:83–97
Yao R et al (2021) Euxanthone inhibits traumatic spinal cord injury via anti-oxidative stress and suppression of p38 and PI3K/Akt signaling pathway in a rat model. Transl Neurosci 12(1):114–126
Zhao R et al (2022) Baicalin attenuates blood-spinal cord barrier disruption and apoptosis through PI3K/Akt signaling pathway after spinal cord injury. Neural Regen Res 17(5):1080–1087
Hu J et al (2021) Chondroitinase ABC Promotes Axon Regeneration and Reduces Retrograde Apoptosis Signaling in Lamprey. Front Cell Dev Biol 9:653638
Li H et al (2020) Initiation of PI3K/AKT pathway by IGF-1 decreases spinal cord injury-induced endothelial apoptosis and microvascular damage. Life Sci 263:118572
Goswami R et al (1999) Overexpression of Akt (protein kinase B) confers protection against apoptosis and prevents formation of ceramide in response to pro-apoptotic stimuli. J Neurosci Res 57(6):884–893
Liao HY et al (2022) Ski regulates proliferation and migration of reactive astrocytes induced by lipopolysaccharide (LPS) through PI3K/Akt pathway. J Neuroimmunol 364:577807
Zhang H, Gong M, Luo X (2020) Methoxytetrahydro-2H-pyran-2-yl)methyl benzoate inhibits spinal cord injury in the rat model via PPAR-γ/PI3K/p-Akt activation. Environ Toxicol 35(6):714–721
Behzadi E, Behzadi P (2016) The role of toll-like receptors (TLRs) in urinary tract infections (UTIs). Cent Eur J Urol 69(4):404–410
Bsibsi M et al (2006) Toll-like receptor 3 on adult human astrocytes triggers production of neuroprotective mediators. Glia 53(7):688–695
Kigerl KA et al (2007) Toll-like receptor (TLR)-2 and TLR-4 regulate inflammation, gliosis, and myelin sparing after spinal cord injury. J Neurochem 102(1):37–50
Lafon M et al (2006) The innate immune facet of brain: human neurons express TLR-3 and sense viral dsRNA. J Mol Neurosci 29(3):185–194
Wang Y et al (2020) Small-Molecule Modulators of Toll-like Receptors. Acc Chem Res 53(5):1046–1055
Akira S, Uematsu S, Takeuchi O (2006) Pathogen recognition and innate immunity. Cell 124(4):783–801
Vaure C, Liu Y (2014) A comparative review of toll-like receptor 4 expression and functionality in different animal species. Front Immunol 5:316
Freria CM et al (2016) Impairment of toll-like receptors 2 and 4 leads to compensatory mechanisms after sciatic nerve axotomy. J Neuroinflammation 13(1):118
Guo X et al (2022) Effects of exosomal microRNAs on oral mucosal epithelial cells cocultured with limbal niche cells. Contrast Media Mol Imaging 2022:9794950
Slota JA, Booth SA (2019) MicroRNAs in neuroinflammation: implications in disease pathogenesis, biomarker discovery and therapeutic applications. Noncoding RNA 5(2):35
Guo Y et al (2019) MicroRNAs in microglia: how do micrornas affect activation, inflammation, polarization of microglia and mediate the interaction between microglia and glioma? Front Mol Neurosci 12:125
Zhou M et al (2018) Abnormal expression of MicroRNAs induced by chronic unpredictable mild stress in rat hippocampal tissues. Mol Neurobiol 55(2):917–935
Liu X et al (2019) MiR-409-3p and MiR-1896 co-operatively participate in IL-17-induced inflammatory cytokine production in astrocytes and pathogenesis of EAE mice via targeting SOCS3/STAT3 signaling. Glia 67(1):101–112
Acknowledgements
We acknowledge the GEO databases for providing their platforms and the contributors for uploading meaningful datasets.
Funding
This work was supported by the Science and Technology Bureau of Quanzhou.
(Grant Number 2022C036R), Medical Innovation Science and Technology Project of Fujian Province (Grant Number 2020CXA047), and Natural Science Foundation of Fujian Province (Grant Number 2020J01227).
Author information
Authors and Affiliations
Contributions
YZ, YSY, and CMW contributed to the study design and data analysis. YZ wrote the paper. WCC and HFH critically revised the manuscript for its intellectual content.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
All animal procedures were performed in accordance with the Care and Use of Laboratory Animals guidelines and were approved by Fujian Medical University.
Consent for Publication
Not applicable.
Competing interests
There author declare no competing conflicts.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Yang, Ys., Chen, Wc. et al. Constructing and Validating a Network of Potential Olfactory Sheathing Cell Transplants Regulating Spinal Cord Injury Progression. Mol Neurobiol 60, 6883–6895 (2023). https://doi.org/10.1007/s12035-023-03510-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-023-03510-9