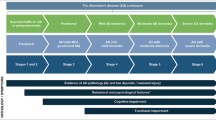
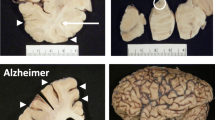
Abstract
Numerous studies have demonstrated an inverse link between cancer and Alzheimer’s disease (AD), with data suggesting that people with Alzheimer’s have a decreased risk of cancer and vice versa. Although other studies have investigated mechanisms to explain this relationship, the connection between these two diseases remains largely unexplained. Processes seen in cancer, such as decreased apoptosis and increased cell proliferation, seem to be reversed in AD. Given the need for effective therapeutic strategies for AD, comparisons with cancer could yield valuable insights into the disease process and perhaps result in new treatments. Here, through a review of existing literature, we compared the expressions of genes involved in cell proliferation and apoptosis to establish a genetic basis for the reciprocal association between AD and cancer. We discuss an array of genes involved in the aforementioned processes, their relevance to both diseases, and how changes in those genes produce varying effects in either disease.

Similar content being viewed by others
Data Availability
This is a review paper. No original data are presented. All data presented are cited with corresponding references and citations.
References
Driver JA, Beiser A, Au R et al (2012) Inverse association between cancer and Alzheimer’s disease: results from the Framingham Heart Study. BMJ 344:e1442. https://doi.org/10.1136/bmj.e1442
Musicco M, Adorni F, Di Santo S et al (2013) Inverse occurrence of cancer and Alzheimer disease: a population-based incidence study. Neurology 81:322–328. https://doi.org/10.1212/WNL.0b013e31829c5ec1
Roe CM, Fitzpatrick AL, Xiong C et al (2010) Cancer linked to Alzheimer disease but not vascular dementia. Neurology 74:106–112. https://doi.org/10.1212/WNL.0b013e3181c91873
Romero JP, Benito-León J, Louis ED, Bermejo-Pareja F (2014) Alzheimer’s disease is associated with decreased risk of cancer-specific mortality: a prospective study (NEDICES). J Alzheimers Dis 40:465–473. https://doi.org/10.3233/JAD-132048
White RS, Lipton RB, Hall CB, Steinerman JR (2013) Nonmelanoma skin cancer is associated with reduced Alzheimer disease risk. Neurology 80:1966–1972. https://doi.org/10.1212/WNL.0b013e3182941990
Zhang Q, Guo S, Zhang X et al (2015) Inverse relationship between cancer and Alzheimer’s disease: a systemic review meta-analysis. Neurol Sci 36:1987–1994. https://doi.org/10.1007/s10072-015-2282-2
Bowles EJA, Walker RL, Anderson ML et al (2017) Risk of Alzheimer’s disease or dementia following a cancer diagnosis. PLoS One 12:e0179857. https://doi.org/10.1371/journal.pone.0179857
Roe CM, Behrens MI, Xiong C et al (2005) Alzheimer disease and cancer. Neurology 64:895–898. https://doi.org/10.1212/01.WNL.0000152889.94785.51
Nudelman KNH, McDonald BC, Lahiri DK, Saykin AJ (2019) Biological hallmarks of cancer in Alzheimer’s disease. Mol Neurobiol 56:7173–7187. https://doi.org/10.1007/s12035-019-1591-5
Schwartz L, Peres S, Jolicoeur M, da Veiga MJ (2020) Cancer and Alzheimer’s disease: intracellular pH scales the metabolic disorders. Biogerontology 21:683–694. https://doi.org/10.1007/s10522-020-09888-6
Xiong Z-G, Pignataro G, Li M et al (2008) Acid-sensing ion channels (ASICs) as pharmacological targets for neurodegenerative diseases. Curr Opin Pharmacol 8:25–32. https://doi.org/10.1016/j.coph.2007.09.001
da Veiga MJ, Peres S, Steyaert J-M et al (2015) Cell cycle progression is regulated by intertwined redox oscillators. Theor Biol Med Model 12:10. https://doi.org/10.1186/s12976-015-0005-2
Paris S, Pouysségur J (1984) Growth factors activate the Na+/H+ antiporter in quiescent fibroblasts by increasing its affinity for intracellular H+. J Biol Chem 259:10989–10994. https://doi.org/10.1016/S0021-9258(18)90611-3
Barker RM, Holly JMP, Biernacka KM et al (2020) Mini review: opposing pathologies in cancer and Alzheimer’s disease: does the PI3K/Akt pathway provide clues? Front Endocrinol 11:403. https://doi.org/10.3389/fendo.2020.00403
Vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029–1033. https://doi.org/10.1126/science.1160809
Zhu X, Perry G, Moreira PI et al (2006) Mitochondrial abnormalities and oxidative imbalance in Alzheimer disease. J Alzheimers Dis 9:147–153. https://doi.org/10.3233/jad-2006-9207
Holohan KN, Lahiri DK, Schneider BP et al (2012) Functional microRNAs in Alzheimer’s disease and cancer: differential regulation of common mechanisms and pathways. Front Genet 3:323. https://doi.org/10.3389/fgene.2012.00323
Driver JA, Zhou XZ, Lu KP (2015) Pin1 dysregulation helps to explain the inverse association between cancer and Alzheimer’s disease. Biochim Biophys Acta 1850:2069–2076. https://doi.org/10.1016/j.bbagen.2014.12.025
Bao L, Kimzey A, Sauter G et al (2004) Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am J Pathol 164:1727–1737. https://doi.org/10.1016/S0002-9440(10)63731-5
Butterfield DA, Abdul HM, Opii W et al (2006) Pin1 in Alzheimer’s disease. J Neurochem 98:1697–1706. https://doi.org/10.1111/j.1471-4159.2006.03995.x
Sultana R, Boyd-Kimball D, Poon HF et al (2006) Oxidative modification and down-regulation of Pin1 in Alzheimer’s disease hippocampus: a redox proteomics analysis. Neurobiol Aging 27:918–925. https://doi.org/10.1016/j.neurobiolaging.2005.05.005
Lu KP, Hanes SD, Hunter T (1996) A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380:544–547. https://doi.org/10.1038/380544a0
Holzer M, Gärtner U, Stöbe A et al (2002) Inverse association of Pin1 and tau accumulation in Alzheimer’s disease hippocampus. Acta Neuropathol 104:471–481. https://doi.org/10.1007/s00401-002-0581-1
SNHG1 small nucleolar RNA host gene 1 [Homo sapiens (human)] - Gene - NCBI. https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=23642. Accessed 2 Aug 2022
Ling J, Yang S, Huang Y et al (2018) Identifying key genes, pathways and screening therapeutic agents for manganese-induced Alzheimer disease using bioinformatics analysis. Medicine 97:e10775. https://doi.org/10.1097/MD.0000000000010775
Gao Y, Zhang N, Lv C et al (2020) lncRNA SNHG1 knockdown alleviates amyloid-β-induced neuronal injury by regulating ZNF217 via sponging miR-361-3p in Alzheimer’s disease. J Alzheimers Dis 77:85–98. https://doi.org/10.3233/JAD-191303
Wang H, Lu B, Chen J (2019) Knockdown of lncRNA SNHG1 attenuated Aβ25-35-inudced neuronal injury via regulating KREMEN1 by acting as a ceRNA of miR-137 in neuronal cells. Biochem Biophys Res Commun 518:438–444. https://doi.org/10.1016/j.bbrc.2019.08.033
Zheng S, Li M, Miao K, Xu H (2019) SNHG1 contributes to proliferation and invasion by regulating miR-382 in breast cancer. Cancer Manag Res 11:5589–5598. https://doi.org/10.2147/CMAR.S198624
Qi H, Wang J, Wang F, Ma H (2017) Long non-coding RNA SNHG1 promotes cell proliferation and tumorigenesis in colorectal cancer via Wnt/β-catenin signaling. Pharmazie 72:395–401. https://doi.org/10.1691/ph.2017.7463
Liu Y, Yang Y, Li L et al (2018) LncRNA SNHG1 enhances cell proliferation, migration, and invasion in cervical cancer. Biochem Cell Biol 96:38–43. https://doi.org/10.1139/bcb-2017-0188
Zhang Y, Jin X, Wang Z et al (2017) Downregulation of SNHG1 suppresses cell proliferation and invasion by regulating Notch signaling pathway in esophageal squamous cell cancer. Cancer Biomark 21:89–96. https://doi.org/10.3233/CBM-170286
Yang Z, Goronzy JJ, Weyand CM (2014) The glycolytic enzyme PFKFB3/phosphofructokinase regulates autophagy. Autophagy 10:382–383. https://doi.org/10.4161/auto.27345
Bolaños JP, Almeida A, Moncada S (2010) Glycolysis: a bioenergetic or a survival pathway? Trends Biochem Sci 35:145–149. https://doi.org/10.1016/j.tibs.2009.10.006
Shi L, Pan H, Liu Z et al (2017) Roles of PFKFB3 in cancer. Signal Transduct Target Ther 2:1–10. https://doi.org/10.1038/sigtrans.2017.44
Herrero-Mendez A, Almeida A, Fernández E et al (2009) The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C–Cdh1. Nat Cell Biol 11:747–752. https://doi.org/10.1038/ncb1881
Almeida A, Moncada S, Bolaños JP (2004) Nitric oxide switches on glycolysis through the AMP protein kinase and 6-phosphofructo-2-kinase pathway. Nat Cell Biol 6:45–51. https://doi.org/10.1038/ncb1080
Fuchsberger T, Martínez-Bellver S, Giraldo E et al (2016) Aβ induces excitotoxicity mediated by APC/C-Cdh1 depletion that can be prevented by glutaminase inhibition promoting neuronal survival. Sci Rep 6:31158. https://doi.org/10.1038/srep31158
Manczak M, Park BS, Jung Y, Reddy PH (2004) Differential expression of oxidative phosphorylation genes in patients with Alzheimer’s disease: implications for early mitochondrial dysfunction and oxidative damage. Neuromolecular Med 5:147–162. https://doi.org/10.1385/NMM:5:2:147
Wang W, Zhao F, Ma X et al (2020) Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Mol Neurodegener 15:30. https://doi.org/10.1186/s13024-020-00376-6
Nagy Z, Esiri MM, LeGris M, Matthews PM (1999) Mitochondrial enzyme expression in the hippocampus in relation to Alzheimer-type pathology. Acta Neuropathol 97:346–354. https://doi.org/10.1007/s004010050997
Ferreira LMR (2010) Cancer metabolism: the Warburg effect today. Exp Mol Pathol 89:372–380. https://doi.org/10.1016/j.yexmp.2010.08.006
Demetrius LA, Magistretti PJ, Pellerin L (2014) Alzheimer’s disease: the amyloid hypothesis and the inverse Warburg effect. Front Physiol 5:522. https://doi.org/10.3389/fphys.2014.00522
Donato R, Cannon BR, Sorci G et al (2013) Functions of S100 proteins. Curr Mol Med 13:24–57. https://doi.org/10.2174/156652413804486214
Breen EC, Tang K (2003) Calcyclin (S100A6) regulates pulmonary fibroblast proliferation, morphology, and cytoskeletal organization in vitro. J Cell Biochem 88:848–854. https://doi.org/10.1002/jcb.10398
Leśniak W, Słomnicki ŁP, Filipek A (2009) S100A6 - new facts and features. Biochem Biophys Res Commun 390:1087–1092. https://doi.org/10.1016/j.bbrc.2009.10.150
Boom A, Pochet R, Authelet M et al (2004) Astrocytic calcium/zinc binding protein S100A6 over expression in Alzheimer’s disease and in PS1/APP transgenic mice models. Biochim Biophys Acta 1742:161–168. https://doi.org/10.1016/j.bbamcr.2004.09.011
Tian Z-Y, Wang C-Y, Wang T et al (2019) Glial S100A6 degrades β-amyloid aggregation through targeting competition with zinc ions. Aging Dis 10:756–769. https://doi.org/10.14336/AD.2018.0912
Yang YQ, Zhang LJ, Dong H et al (2007) Upregulated expression of S100A6 in human gastric cancer. J Dig Dis 8:186–193. https://doi.org/10.1111/j.1751-2980.2007.00311.x
Wang X-H, Du H, Li L et al (2017) Increased expression of S100A6 promotes cell proliferation in gastric cancer cells. Oncol Lett 13:222–230. https://doi.org/10.3892/ol.2016.5419
Hua Z, Chen J, Sun B et al (2011) Specific expression of osteopontin and S100A6 in hepatocellular carcinoma. Surgery 149:783–791. https://doi.org/10.1016/j.surg.2010.12.007
Li A, Gu Y, Li X et al (2018) S100A6 promotes the proliferation and migration of cervical cancer cells via the PI3K/Akt signaling pathway. Oncol Lett 15:5685–5693. https://doi.org/10.3892/ol.2018.8018
Turner NC, Reis-Filho JS (2006) Basal-like breast cancer and the BRCA1 phenotype. Oncogene 25:5846–5853. https://doi.org/10.1038/sj.onc.1209876
Wang Z, Zhang J, Zhang Y et al (2018) Expression and mutations of BRCA in breast cancer and ovarian cancer: evidence from bioinformatics analyses. Int J Mol Med 42:3542–3550. https://doi.org/10.3892/ijmm.2018.3870
Shan J, Dsouza SP, Bakhru S et al (2013) TNRC9 downregulates BRCA1 expression and promotes breast cancer aggressiveness. Cancer Res 73:2840–2849. https://doi.org/10.1158/0008-5472.CAN-12-4313
Zhang W, Luo J, Yang F et al (2016) BRCA1 inhibits AR-mediated proliferation of breast cancer cells through the activation of SIRT1. Sci Rep 6:22034. https://doi.org/10.1038/srep22034
Yoshida K, Miki Y (2004) Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci 95:866–871. https://doi.org/10.1111/j.1349-7006.2004.tb02195.x
Gudas JM, Li T, Nguyen H et al (1996) Cell cycle regulation of BRCA1 messenger RNA in human breast epithelial cells. Cell Growth Differ 7:717–723
Thompson ME, Jensen RA, Obermiller PS et al (1995) Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat Genet 9:444–450. https://doi.org/10.1038/ng0495-444
Suberbielle E, Sanchez PE, Kravitz AV et al (2013) Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat Neurosci 16:613–621. https://doi.org/10.1038/nn.3356
Mano T, Nagata K, Nonaka T et al (2017) Neuron-specific methylome analysis reveals epigenetic regulation and tau-related dysfunction of BRCA1 in Alzheimer’s disease. Proc Natl Acad Sci U S A 114:E9645–E9654. https://doi.org/10.1073/pnas.1707151114
Wezyk M, Zekanowski C (2018) Role of BRCA1 in neuronal death in Alzheimer’s disease. ACS Chem Neurosci 9:870–872. https://doi.org/10.1021/acschemneuro.8b00149
Wezyk M, Szybinska A, Wojsiat J et al (2018) Overactive BRCA1 affects presenilin 1 in induced pluripotent stem cell-derived neurons in Alzheimer’s disease. J Alzheimers Dis 62:175–202. https://doi.org/10.3233/JAD-170830
Welch MD, DePace AH, Verma S et al (1997) The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol 138:375–384. https://doi.org/10.1083/jcb.138.2.375
Lauterborn JC, Cox CD, Chan SW et al (2020) Synaptic actin stabilization protein loss in down syndrome and Alzheimer disease. Brain Pathol 30:319–331. https://doi.org/10.1111/bpa.12779
Rauhala HE, Teppo S, Niemelä S, Kallioniemi A (2013) Silencing of the ARP2/3 complex disturbs pancreatic cancer cell migration. Anticancer Res 33:45–52
DiBattista AM, Dumanis SB, Song JM et al (2015) Very low density lipoprotein receptor regulates dendritic spine formation in a RasGRF1/CaMKII dependent manner. Biochim Biophys Acta 1853:904–917. https://doi.org/10.1016/j.bbamcr.2015.01.015
Deutsch SI, Rosse RB, Deutsch LH (2006) Faulty regulation of tau phosphorylation by the reelin signal transduction pathway is a potential mechanism of pathogenesis and therapeutic target in Alzheimer’s disease. Eur Neuropsychopharmacol 16:547–551. https://doi.org/10.1016/j.euroneuro.2006.01.006
Hiesberger T, Trommsdorff M, Howell BW et al (1999) Direct binding of Reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron 24:481–489. https://doi.org/10.1016/s0896-6273(00)80861-2
Zhou H, Guo W, Zhao Y et al (2014) MicroRNA-135a acts as a putative tumor suppressor by directly targeting very low density lipoprotein receptor in human gallbladder cancer. Cancer Sci 105:956–965. https://doi.org/10.1111/cas.12463
He L, Lu Y, Guo J (2013) Type II VLDLR promotes cell migration by up-regulation of VEGF, MMP2 and MMP7 in breast cancer cells. Chin Ger J Clin Oncol 12:374–378. https://doi.org/10.1007/s10330-013-1218-7
Sanchez-Mut JV, Aso E, Panayotis N et al (2013) DNA methylation map of mouse and human brain identifies target genes in Alzheimer’s disease. Brain 136:3018–3027. https://doi.org/10.1093/brain/awt237
Astarita G, Jung K-M, Vasilevko V et al (2011) Elevated stearoyl-CoA desaturase in brains of patients with Alzheimer’s disease. PLoS One 6:e24777. https://doi.org/10.1371/journal.pone.0024777
Hu W, Lin X, Chen K (2015) Integrated analysis of differential gene expression profiles in hippocampi to identify candidate genes involved in Alzheimer’s disease. Mol Med Rep 12:6679–6687. https://doi.org/10.3892/mmr.2015.4271
Ma SL, Tang NLS, Lam LCW (2016) Association of gene expression and methylation of UQCRC1 to the predisposition of Alzheimer’s disease in a Chinese population. J Psychiatr Res 76:143–147. https://doi.org/10.1016/j.jpsychires.2016.02.010
Li X, Long J, He T et al (2015) Integrated genomic approaches identify major pathways and upstream regulators in late onset Alzheimer’s disease. Sci Rep 5:1–12. https://doi.org/10.1038/srep12393
Cagliani R, Guerini FR, Rubio-Acero R et al (2013) Long-standing balancing selection in the THBS4 gene: influence on sex-specific brain expression and gray matter volumes in Alzheimer disease. Hum Mutat 34:743–753. https://doi.org/10.1002/humu.22301
Strachan GD, Ostrow LA, Jordan-Sciutto KL (2005) Expression of the fetal Alz-50 clone 1 protein induces apoptotic cell death. Biochem Biophys Res Commun 336:490–495. https://doi.org/10.1016/j.bbrc.2005.08.127
Carter CJ (2007) Convergence of genes implicated in Alzheimer’s disease on the cerebral cholesterol shuttle: APP, cholesterol, lipoproteins, and atherosclerosis. Neurochem Int 50:12–38. https://doi.org/10.1016/j.neuint.2006.07.007
Lu W, Mi R, Tang H et al (1998) Over-expression of c-fos mRNA in the hippocampal neurons in Alzheimer’s disease. Chin Med J 111:35–37
Cardona K, Medina J, Orrego-Cardozo M et al (2021) Inflammatory gene expression profiling in peripheral blood from patients with Alzheimer’s disease reveals key pathways and hub genes with potential diagnostic utility: a preliminary study. PeerJ 9:e12016. https://doi.org/10.7717/peerj.12016
de la Monte SM, Wands JR (2006) Molecular indices of oxidative stress and mitochondrial dysfunction occur early and often progress with severity of Alzheimer’s disease. J Alzheimers Dis 9:167–181. https://doi.org/10.3233/jad-2006-9209
Tian T, Qiu R, Qiu X (2018) SNHG1 promotes cell proliferation by acting as a sponge of miR-145 in colorectal cancer. Oncotarget 9:2128–2139. https://doi.org/10.18632/oncotarget.23255
Ploeger C, Waldburger N, Fraas A et al (2016) Chromosome 8p tumor suppressor genes SH2D4A and SORBS3 cooperate to inhibit interleukin-6 signaling in hepatocellular carcinoma. Hepatology 64:828–842. https://doi.org/10.1002/hep.28684
Kim S-J, Choi H, Park S-S et al (2011) Stearoyl CoA desaturase (SCD) facilitates proliferation of prostate cancer cells through enhancement of androgen receptor transactivation. Mol Cells 31:371–377. https://doi.org/10.1007/s10059-011-0043-5
Zhao X, Chen M, Tan J (2016) Knockdown of ZFR suppresses cell proliferation and invasion of human pancreatic cancer. Biol Res 49:26. https://doi.org/10.1186/s40659-016-0086-3
Han J, Meng Q, Xi Q et al (2017) PFKFB3 was overexpressed in gastric cancer patients and promoted the proliferation and migration of gastric cancer cells. Cancer Biomark 18:249–256. https://doi.org/10.3233/CBM-160143
Wang X-H, Zhang L-H, Zhong X-Y et al (2010) S100A6 overexpression is associated with poor prognosis and is epigenetically up-regulated in gastric cancer. Am J Pathol 177:586–597. https://doi.org/10.2353/ajpath.2010.091217
Chen B, Huang T, Jiang J et al (2014) miR-141 suppresses proliferation and motility of gastric cancer cells by targeting HDGF. Mol Cell Biochem 388:211–218. https://doi.org/10.1007/s11010-013-1912-3
Wang Q, Li M, Gan Y et al (2020) Mitochondrial protein UQCRC1 is oncogenic and a potential therapeutic target for pancreatic cancer. Theranostics 10:2141–2157. https://doi.org/10.7150/thno.38704
Egusquiaguirre SP, Yeh JE, Walker SR et al (2018) The STAT3 target gene TNFRSF1A modulates the NF-κB pathway in breast cancer cells. Neoplasia 20:489–498. https://doi.org/10.1016/j.neo.2018.03.004
Greco SA, Chia J, Inglis KJ et al (2010) Thrombospondin-4 is a putative tumour-suppressor gene in colorectal cancer that exhibits age-related methylation. BMC Cancer 10:494. https://doi.org/10.1186/1471-2407-10-494
Dai M, Lu J-J, Guo W et al (2015) BPTF promotes tumor growth and predicts poor prognosis in lung adenocarcinomas. Oncotarget 6:33878–33892. https://doi.org/10.18632/oncotarget.5302
Yang G-D, Yang X-M, Lu H et al (2014) SERPINA3 promotes endometrial cancer cells growth by regulating G2/M cell cycle checkpoint and apoptosis. Int J Clin Exp Pathol 7:1348–1358
Yuspa SH (1998) The pathogenesis of squamous cell cancer: lessons learned from studies of skin carcinogenesis. J Dermatol Sci 17:1–7. https://doi.org/10.1016/s0923-1811(97)00071-6
Ding L, Xu Y, Zhang W et al (2010) MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res 20:784–793. https://doi.org/10.1038/cr.2010.79
Zou Z, Li X, Sun Y et al (2020) NOS1 expression promotes proliferation and invasion and enhances chemoresistance in ovarian cancer. Oncol Lett 19:2989–2995. https://doi.org/10.3892/ol.2020.11355
Mirza Z, Rajeh N (2017) Identification of electrophysiological changes in Alzheimer’s disease: a microarray based transcriptomics and molecular pathway analysis study. CNS Neurol Disord Drug Targets 16:1027–1038. https://doi.org/10.2174/1871527316666171023153837
Wilmot B, McWeeney SK, Nixon RR et al (2008) Translational gene mapping of cognitive decline. Neurobiol Aging 29:524–541. https://doi.org/10.1016/j.neurobiolaging.2006.11.008
Taguchi K, Yamagata HD, Zhong W et al (2005) Identification of hippocampus-related candidate genes for Alzheimer’s disease. Ann Neurol 57:585–588. https://doi.org/10.1002/ana.20433
Annese A, Manzari C, Lionetti C et al (2018) Whole transcriptome profiling of late-onset Alzheimer’s disease patients provides insights into the molecular changes involved in the disease. Sci Rep 8:1–15. https://doi.org/10.1038/s41598-018-22701-2
Castillo E, Leon J, Mazzei G et al (2017) Comparative profiling of cortical gene expression in Alzheimer’s disease patients and mouse models demonstrates a link between amyloidosis and neuroinflammation. Sci Rep 7:17762. https://doi.org/10.1038/s41598-017-17999-3
Lv L, Zhang D, Hua P, Yang S (2021) The glial-specific hypermethylated 3’ untranslated region of histone deacetylase 1 may modulates several signal pathways in Alzheimer’s disease. Life Sci 265:118760. https://doi.org/10.1016/j.lfs.2020.118760
Herring A, Donath A, Steiner KM et al (2012) Reelin depletion is an early phenomenon of Alzheimer’s pathology. J Alzheimers Dis 30:963–979. https://doi.org/10.3233/JAD-2012-112069
Talwar P, Silla Y, Grover S et al (2014) Genomic convergence and network analysis approach to identify candidate genes in Alzheimer’s disease. BMC Genomics 15:199. https://doi.org/10.1186/1471-2164-15-199
Gómez Ravetti M, Rosso OA, Berretta R, Moscato P (2010) Uncovering molecular biomarkers that correlate cognitive decline with the changes of hippocampus’ gene expression profiles in Alzheimer’s disease. PLoS One 5:e10153. https://doi.org/10.1371/journal.pone.0010153
Bennett JP, Keeney PM (2017) Micro RNA’s (mirna’s) may help explain expression of multiple genes in Alzheimer’s Frontal Cortex. J Syst Integr Neurosci 3:1–9. https://doi.org/10.15761/jsin.1000178
Acquaah-Mensah GK, Agu N, Khan T, Gardner A (2015) A regulatory role for the insulin- and BDNF-linked RORA in the hippocampus: implications for Alzheimer’s disease. J Alzheimers Dis 44:827–838. https://doi.org/10.3233/JAD-141731
Liu X, Jiao B, Shen L (2018) The epigenetics of Alzheimer’s disease: factors and therapeutic implications. Front Genet 9:579. https://doi.org/10.3389/fgene.2018.00579
Barbash S, Soreq H (2012) Threshold-independent meta-analysis of Alzheimer’s disease transcriptomes shows progressive changes in hippocampal functions, epigenetics and microRNA regulation. Curr Alzheimer Res 9:425–435. https://doi.org/10.2174/156720512800492512
Liang WS, Dunckley T, Beach TG et al (2008) Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol Genomics 33:240–256. https://doi.org/10.1152/physiolgenomics.00242.2007
Wang Y, Zheng T (2014) Screening of hub genes and pathways in colorectal cancer with microarray technology. Pathol Oncol Res 20:611–618. https://doi.org/10.1007/s12253-013-9739-5
Yu B-L, Peng X-H, Zhao F-P et al (2014) MicroRNA-378 functions as an onco-miR in nasopharyngeal carcinoma by repressing TOB2 expression. Int J Oncol 44:1215–1222. https://doi.org/10.3892/ijo.2014.2283
Xie C, Xiong W, Li J et al (2019) Intersectin 1 (ITSN1) identified by comprehensive bioinformatic analysis and experimental validation as a key candidate biological target in breast cancer. Onco Targets Ther 12:7079–7093. https://doi.org/10.2147/OTT.S216286
Shen L, Yang M, Lin Q et al (2016) COL11A1 is overexpressed in recurrent non-small cell lung cancer and promotes cell proliferation, migration, invasion and drug resistance. Oncol Rep 36:877–885. https://doi.org/10.3892/or.2016.4869
Xu N, Qu G-Y, Wu Y-P et al (2020) ARPC4 promotes bladder cancer cell invasion and is associated with lymph node metastasis. J Cell Biochem 121:231–243. https://doi.org/10.1002/jcb.29136
Liu Z, Gu S, Lu T et al (2020) IFI6 depletion inhibits esophageal squamous cell carcinoma progression through reactive oxygen species accumulation via mitochondrial dysfunction and endoplasmic reticulum stress. J Exp Clin Cancer Res 39:144. https://doi.org/10.1186/s13046-020-01646-3
Gröschl B, Bettstetter M, Giedl C et al (2013) Expression of the MAP kinase phosphatase DUSP4 is associated with microsatellite instability in colorectal cancer (CRC) and causes increased cell proliferation. Int J Cancer 132:1537–1546. https://doi.org/10.1002/ijc.27834
Chen Y, Song Y, Wang Z et al (2010) Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J Gastrointest Surg 14:1170–1179. https://doi.org/10.1007/s11605-010-1202-2
Xia Y, Gao Y (2014) MicroRNA-181b promotes ovarian cancer cell growth and invasion by targeting LATS2. Biochem Biophys Res Commun 447:446–451. https://doi.org/10.1016/j.bbrc.2014.04.027
He L, Lu Y, Wang P et al (2010) Up-regulated expression of type II very low density lipoprotein receptor correlates with cancer metastasis and has a potential link to β-catenin in different cancers. BMC Cancer 10:601. https://doi.org/10.1186/1471-2407-10-601
Bi D-P, Yin C-H, Zhang X-Y et al (2016) MiR-183 functions as an oncogene by targeting ABCA1 in colon cancer. Oncol Rep 35:2873–2879. https://doi.org/10.3892/or.2016.4631
Lu Y, Ma J, Li Y et al (2017) CDP138 silencing inhibits TGF-β/Smad signaling to impair radioresistance and metastasis via GDF15 in lung cancer. Cell Death Dis 8:e3036–e3036. https://doi.org/10.1038/cddis.2017.434
Li J, Feng X, Sun C et al (2015) Associations between proteasomal activator PA28γ and outcome of oral squamous cell carcinoma: evidence from cohort studies and functional analyses. EBioMedicine 2:851–858. https://doi.org/10.1016/j.ebiom.2015.07.004
Xu X-W, Yang X-M, Zhao W-J et al (2018) DNM1L, a key prognostic predictor for gastric adenocarcinoma, is involved in cell proliferation, invasion, and apoptosis. Oncol Lett 16:3635–3641. https://doi.org/10.3892/ol.2018.9138
Dobrovic A, Simpfendorfer D (1997) Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res 57:3347–3350
Zhao Y, Wang X, Wang T et al (2011) Acetylcholinesterase, a key prognostic predictor for hepatocellular carcinoma, suppresses cell growth and induces chemosensitization. Hepatology 53:493–503. https://doi.org/10.1002/hep.24079
Guo Y, Bao Y, Ma M et al (2017) Clinical significance of the correlation between PLCE 1 and PRKCA in esophageal inflammation and esophageal carcinoma. Oncotarget 8:33285–33299. https://doi.org/10.18632/oncotarget.16635
Asamitsu K, Tetsuka T, Kanazawa S, Okamoto T (2003) RING finger protein AO7 supports NF-kappaB-mediated transcription by interacting with the transactivation domain of the p65 subunit. J Biol Chem 278:26879–26887. https://doi.org/10.1074/jbc.M211831200
Yamaguchi Y, Ayaki T, Li F et al (2019) Phosphorylated NF-κB subunit p65 aggregates in granulovacuolar degeneration and neurites in neurodegenerative diseases with tauopathy. Neurosci Lett 704:229–235. https://doi.org/10.1016/j.neulet.2019.03.036
Valerio A, Boroni F, Benarese M et al (2006) NF-kappaB pathway: a target for preventing beta-amyloid (Abeta)-induced neuronal damage and Abeta42 production. Eur J Neurosci 23:1711–1720. https://doi.org/10.1111/j.1460-9568.2006.04722.x
Lin W, Ding M, Xue J, Leng W (2013) The role of TLR2/JNK/NF-κB pathway in amyloid β peptide-induced inflammatory response in mouse NG108-15 neural cells. Int Immunopharmacol 17:880–884. https://doi.org/10.1016/j.intimp.2013.09.016
Lindsay A, Hickman D, Srinivasan M (2021) A nuclear factor-kappa B inhibiting peptide suppresses innate immune receptors and gliosis in a transgenic mouse model of Alzheimer’s disease. Biomed Pharmacother 138:111405. https://doi.org/10.1016/j.biopha.2021.111405
Yu H-G, Yu L-L, Yang Y et al (2003) Increased expression of RelA/nuclear factor-kappa B protein correlates with colorectal tumorigenesis. Oncology 65:37–45. https://doi.org/10.1159/000071203
Sovak MA, Bellas RE, Kim DW et al (1997) Aberrant nuclear factor-kappaB/Rel expression and the pathogenesis of breast cancer. J Clin Invest 100:2952–2960. https://doi.org/10.1172/JCI119848
Tai DI, Tsai SL, Chang YH et al (2000) Constitutive activation of nuclear factor kappaB in hepatocellular carcinoma. Cancer 89:2274–2281
Olbrich P, Freeman AF (2018) STAT1 and STAT3 mutations: important lessons for clinical immunologists. Expert Rev Clin Immunol 14:1029–1041. https://doi.org/10.1080/1744666X.2018.1531704
Lu Y, Li K, Hu Y, Wang X (2021) Expression of immune related genes and possible regulatory mechanisms in Alzheimer’s disease. Front Immunol 12:768966. https://doi.org/10.3389/fimmu.2021.768966
Reichenbach N, Delekate A, Plescher M et al (2019) Inhibition of Stat3-mediated astrogliosis ameliorates pathology in an Alzheimer’s disease model. EMBO Mol Med 11:e9665. https://doi.org/10.15252/emmm.201809665
Ceyzériat K, Ben Haim L, Denizot A et al (2018) Modulation of astrocyte reactivity improves functional deficits in mouse models of Alzheimer’s disease. Acta Neuropathol Commun 6:104. https://doi.org/10.1186/s40478-018-0606-1
Wan J, Fu AKY, Ip FCF et al (2010) Tyk2/STAT3 signaling mediates beta-amyloid-induced neuronal cell death: implications in Alzheimer’s disease. J Neurosci 30:6873–6881. https://doi.org/10.1523/JNEUROSCI.0519-10.2010
Cui X, Jing X, Yi Q et al (2018) Systematic analysis of gene expression alterations and clinical outcomes of STAT3 in cancer. Oncotarget 9:3198–3213. https://doi.org/10.18632/oncotarget.23226
Wang Y, Shen Y, Wang S et al (2018) The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett 415:117–128. https://doi.org/10.1016/j.canlet.2017.12.003
Yuan F, Fu X, Shi H et al (2014) Induction of murine macrophage M2 polarization by cigarette smoke extract via the JAK2/STAT3 pathway. PLoS One 9:e107063. https://doi.org/10.1371/journal.pone.0107063
Rahaman SO, Harbor PC, Chernova O et al (2002) Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene 21:8404–8413. https://doi.org/10.1038/sj.onc.1206047
Maes ME, Schlamp CL, Nickells RW (2017) BAX to basics: how the BCL2 gene family controls the death of retinal ganglion cells. Prog Retin Eye Res 57:1–25. https://doi.org/10.1016/j.preteyeres.2017.01.002
O’Barr S, Schultz J, Rogers J (1996) Expression of the protooncogene bcl-2 in Alzheimer’s disease brain. Neurobiol Aging 17:131–136. https://doi.org/10.1016/0197-4580(95)02024-1
Webster S, O’Barr S, Rogers J (1994) Enhanced aggregation and beta structure of amyloid beta peptide after coincubation with C1q. J Neurosci Res 39:448–456. https://doi.org/10.1002/jnr.490390412
Thomadaki H, Talieri M, Scorilas A (2007) Prognostic value of the apoptosis related genes BCL2 and BCL2L12 in breast cancer. Cancer Lett 247:48–55. https://doi.org/10.1016/j.canlet.2006.03.016
Ghaffari M, Kalantar SM, Hemati M et al (2021) Co-delivery of miRNA-15a and miRNA-16-1 using cationic PEGylated niosomes downregulates Bcl-2 and induces apoptosis in prostate cancer cells. Biotechnol Lett 43:981–994. https://doi.org/10.1007/s10529-021-03085-2
Karch CM, Cruchaga C, Goate AM (2014) Alzheimer’s disease genetics: from the bench to the clinic. Neuron 83:11–26. https://doi.org/10.1016/j.neuron.2014.05.041
Del Villar K, Miller CA (2004) Down-regulation of DENN/MADD, a TNF receptor binding protein, correlates with neuronal cell death in Alzheimer’s disease brain and hippocampal neurons. Proc Natl Acad Sci U S A 101:4210–4215. https://doi.org/10.1073/pnas.0307349101
Miyoshi J, Takai Y (2004) Dual role of DENN/MADD (Rab3GEP) in neurotransmission and neuroprotection. Trends Mol Med 10:476–480. https://doi.org/10.1016/j.molmed.2004.08.002
Del Villar K, Miller CA (2003) Oxidative stress and death domain proteins in Alzheimer’s disease. Clin Neurosci Res 2:316–323. https://doi.org/10.1016/S1566-2772(03)00008-2
Li L-C, Jayaram S, Ganesh L et al (2011) Knockdown of MADD and c-FLIP overcomes resistance to TRAIL-induced apoptosis in ovarian cancer cells. Am J Obstet Gynecol 205:362.e12–25. https://doi.org/10.1016/j.ajog.2011.05.035
Turner A, Li L-C, Pilli T et al (2013) MADD knock-down enhances doxorubicin and TRAIL induced apoptosis in breast cancer cells. PLoS One 8:e56817. https://doi.org/10.1371/journal.pone.0056817
Saini S, Sripada L, Tulla K et al (2019) Loss of MADD expression inhibits cellular growth and metastasis in anaplastic thyroid cancer. Cell Death Dis 10:1–13. https://doi.org/10.1038/s41419-019-1351-5
DAB1 DAB adaptor protein 1 [Homo sapiens (human)] - Gene - NCBI. https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=1600. Accessed 2 Aug 2022
Müller T, Loosse C, Schrötter A et al (2011) The AICD interacting protein DAB1 is up-regulated in Alzheimer frontal cortex brain samples and causes deregulation of proteins involved in gene expression changes. Curr Alzheimer Res 8:573–582. https://doi.org/10.2174/156720511796391827
McAvoy S, Zhu Y, Perez DS et al (2008) Disabled-1 is a large common fragile site gene, inactivated in multiple cancers. Genes Chromosomes Cancer 47:165–174. https://doi.org/10.1002/gcc.20519
Li L, Hao J, Yan C-Q et al (2020) Inhibition of microRNA-300 inhibits cell adhesion, migration, and invasion of prostate cancer cells by promoting the expression of DAB1. Cell Cycle 19:2793–2810. https://doi.org/10.1080/15384101.2020.1823730
Cao R-J, Li K, Xing W-Y et al (2019) Disabled-1 is down-regulated in clinical breast cancer and regulates cell apoptosis through NF-κB/Bcl-2/caspase-9. J Cell Mol Med 23:1622–1627. https://doi.org/10.1111/jcmm.14047
Rohn TT, Rissman RA, Davis MC et al (2002) Caspase-9 activation and caspase cleavage of tau in the Alzheimer’s disease brain. Neurobiol Dis 11:341–354. https://doi.org/10.1006/nbdi.2002.0549
Tamayev R, Akpan N, Arancio O et al (2012) Caspase-9 mediates synaptic plasticity and memory deficits of Danish dementia knock-in mice: caspase-9 inhibition provides therapeutic protection. Mol Neurodegener 7:60. https://doi.org/10.1186/1750-1326-7-60
Boghaert ER, Sells SF, Walid AJ et al (1997) Immunohistochemical analysis of the proapoptotic protein Par-4 in normal rat tissues. Cell Growth Differ 8:881–890
Xie J, Guo Q (2005) PAR-4 is involved in regulation of beta-secretase cleavage of the Alzheimer amyloid precursor protein. J Biol Chem 280:13824–13832. https://doi.org/10.1074/jbc.M411933200
Guo Q, Fu W, Xie J et al (1998) Par-4 is a mediator of neuronal degeneration associated with the pathogenesis of Alzheimer disease. Nat Med 4:957–962. https://doi.org/10.1038/nm0898-957
Xie J, Chang X, Zhang X, Guo Q (2001) Aberrant induction of Par-4 is involved in apoptosis of hippocampal neurons in presenilin-1 M146V mutant knock-in mice. Brain Res 915:1–10. https://doi.org/10.1016/s0006-8993(01)02803-7
Guo Q, Xie J, Chang X et al (2001) Par-4 is a synaptic protein that regulates neurite outgrowth by altering calcium homeostasis and transcription factor AP-1 activation. Brain Res 903:13–25. https://doi.org/10.1016/s0006-8993(01)02304-6
Xie J, Awad KS, Guo Q (2005) RNAi knockdown of Par-4 inhibits neurosynaptic degeneration in ALS-linked mice. J Neurochem 92:59–71. https://doi.org/10.1111/j.1471-4159.2004.02834.x
Lu D, Tang L, Zhuang Y, Zhao P (2018) miR-17-3P regulates the proliferation and survival of colon cancer cells by targeting Par4. Mol Med Rep 17:618–623. https://doi.org/10.3892/mmr.2017.7863
Watson CN, Begum G, Ashman E et al (2022) Co-expression analysis of microRNAs and proteins in brain of Alzheimer’s disease patients. Cells 11:163. https://doi.org/10.3390/cells11010163
Ascolani A, Balestrieri E, Minutolo A et al (2012) Dysregulated NF-κB pathway in peripheral mononuclear cells of Alzheimer’s disease patients. Curr Alzheimer Res 9:128–137. https://doi.org/10.2174/156720512799015091
Kitamura Y, Shimohama S, Ota T et al (1997) Alteration of transcription factors NF-kappaB and STAT1 in Alzheimer’s disease brains. Neurosci Lett 237:17–20. https://doi.org/10.1016/s0304-3940(97)00797-0
Ye M-F, Lin D, Li W-J et al (2020) MiR-26a-5p serves as an oncogenic microRNA in non-small cell lung cancer by targeting FAF1. Cancer Manag Res 12:7131–7142. https://doi.org/10.2147/CMAR.S261131
Wang Q, Gao G, Zhang T et al (2018) TRAF1 is critical for regulating the BRAF/MEK/ERK pathway in non-small cell lung carcinogenesis. Cancer Res 78:3982–3994. https://doi.org/10.1158/0008-5472.CAN-18-0429
Weichert W, Boehm M, Gekeler V et al (2007) High expression of RelA/p65 is associated with activation of nuclear factor-κB-dependent signaling in pancreatic cancer and marks a patient population with poor prognosis. Br J Cancer 97:523–530. https://doi.org/10.1038/sj.bjc.6603878
Don-Doncow N, Marginean F, Coleman I et al (2017) Expression of STAT3 in prostate cancer metastases. Eur Urol 71:313–316. https://doi.org/10.1016/j.eururo.2016.06.018
Tosto G, Fu H, Vardarajan BN et al (2015) F-box/LRR-repeat protein 7 is genetically associated with Alzheimer’s disease. Ann Clin Transl Neurol 2:810–820. https://doi.org/10.1002/acn3.223
Sajan FD, Martiniuk F, Marcus DL et al (2007) Apoptotic gene expression in Alzheimer’s disease hippocampal tissue. Am J Alzheimers Dis Other Demen 22:319–328. https://doi.org/10.1177/1533317507302447
Peng Y-S, Tang C-W, Peng Y-Y et al (2020) Comparative functional genomic analysis of Alzheimer’s affected and naturally aging brains. PeerJ 8:e8682. https://doi.org/10.7717/peerj.8682
Wang X, Michaelis ML, Michaelis EK (2010) Functional genomics of brain aging and Alzheimer’s disease: focus on selective neuronal vulnerability. Curr Genomics 11:618–633. https://doi.org/10.2174/138920210793360943
González P, Alvarez V, Menéndez M et al (2007) Myocyte enhancing factor-2A in Alzheimer’s disease: genetic analysis and association with MEF2A-polymorphisms. Neurosci Lett 411:47–51. https://doi.org/10.1016/j.neulet.2006.09.055
McKeever PM, Kim T, Hesketh AR et al (2017) Cholinergic neuron gene expression differences captured by translational profiling in a mouse model of Alzheimer’s disease. Neurobiol Aging 57:104–119. https://doi.org/10.1016/j.neurobiolaging.2017.05.014
Hague A, Moorghen M, Hicks D et al (1994) BCL-2 expression in human colorectal adenomas and carcinomas. Oncogene 9:3367–3370
Moro L, Pagano M (2020) Epigenetic suppression of FBXL7 promotes metastasis. Mol Cell Oncol 7:1833698. https://doi.org/10.1080/23723556.2020.1833698
Valladares A, Hernández NG, Gómez FS et al (2006) Genetic expression profiles and chromosomal alterations in sporadic breast cancer in Mexican women. Cancer Genet Cytogenet 170:147–151. https://doi.org/10.1016/j.cancergencyto.2006.06.002
Bi W, Wei Y, Wu J et al (2013) MADD promotes the survival of human lung adenocarcinoma cells by inhibiting apoptosis. Oncol Rep 29:1533–1539. https://doi.org/10.3892/or.2013.2258
Cai M-J, Cui Y, Fang M et al (2019) Inhibition of PSMD4 blocks the tumorigenesis of hepatocellular carcinoma. Gene 702:66–74. https://doi.org/10.1016/j.gene.2019.03.063
Dunn TA, Chen S, Faith DA et al (2006) A novel role of myosin VI in human prostate cancer. Am J Pathol 169:1843–1854. https://doi.org/10.2353/ajpath.2006.060316
Sato N, Fukushima N, Chang R et al (2006) Differential and epigenetic gene expression profiling identifies frequent disruption of the RELN pathway in pancreatic cancers. Gastroenterology 130:548–565. https://doi.org/10.1053/j.gastro.2005.11.008
Zhang Y, Yu G, Jiang P et al (2011) Decreased expression of protease-activated receptor 4 in human gastric cancer. Int J Biochem Cell Biol 43:1277–1283. https://doi.org/10.1016/j.biocel.2011.05.008
Bai X, Wu L, Liang T et al (2008) Overexpression of myocyte enhancer factor 2 and histone hyperacetylation in hepatocellular carcinoma. J Cancer Res Clin Oncol 134:83–91. https://doi.org/10.1007/s00432-007-0252-7
Kusy S, Potiron V, Zeng C et al (2005) Promoter characterization of Semaphorin SEMA3F, a tumor suppressor gene. Biochim Biophys Acta 1730:66–76. https://doi.org/10.1016/j.bbaexp.2005.05.008
Ospina-Romero M, Abdiwahab E, Kobayashi L et al (2019) Rate of memory change before and after cancer diagnosis. JAMA Netw Open 2:e196160. https://doi.org/10.1001/jamanetworkopen.2019.6160
Acknowledgements
The authors acknowledge the financial support of the Krembil Foundation.
Funding
This work was supported by an operating grant from the Krembil Foundation.
Author information
Authors and Affiliations
Contributions
All authors contributed to the development and writing of this review. The first draft of the manuscript was written by Aditya Bhardwaj and Imindu Liyanage. D. Weaver contributed to all subsequent drafts. All authors commented on all successive versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
This is a review paper. The UHN Research Ethics Committee has confirmed that no ethical approval is required.
Consent to Participate
There was no research involving human subjects.
Consent for Publication
Consent to publish is not required.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bhardwaj, A., Liyanage, S.I. & Weaver, D.F. Cancer and Alzheimer’s Inverse Correlation: an Immunogenetic Analysis. Mol Neurobiol 60, 3086–3099 (2023). https://doi.org/10.1007/s12035-023-03260-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-023-03260-8