Abstract
Hemiplegic migraine (HM) is a rare subtype of migraine with aura. Given that causal missense mutations in the voltage-gated calcium channel α1A subunit gene CACNA1A have been identified in a subset of HM patients, we investigated whether HM patients without a mutation have an increased burden of such variants in the “CACNA1x gene family”. Whole exome sequencing data of an Australian cohort of unrelated HM patients (n = 184), along with public data from gnomAD, as controls, was used to assess the burden of missense variants in CACNA1x genes. We performed both a variant and a subject burden test. We found a significant burden for the number of variants in CACNA1E (p = 1.3 × 10−4), CACNA1H (p < 2.2 × 10−16) and CACNA1I (p < 2.2 × 10−16). There was also a significant burden of subjects with missense variants in CACNA1E (p = 6.2 × 10−3), CACNA1H (p < 2.2 × 10−16) and CACNA1I (p < 2.2 × 10−16). Both the number of variants and number of subjects were replicated for CACNA1H (p = 3.5 × 10−8; p = 0.012) and CACNA1I (p = 0.019, p = 0.044), respectively, in a Dutch clinical HM cohort (n = 32), albeit that CACNA1I did not remain significant after multiple testing correction. Our data suggest that HM, in the absence of a single causal mutation, is a complex trait, in which an increased burden of missense variants in CACNA1H and CACNA1I may contribute to the risk of disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Hemiplegic migraine (HM) is a rare subtype of migraine with aura with attacks that are associated with motor weakness or hemiplegia during the aura phase [1]. HM is clinically and genetically heterogeneous [2,3,4] and can be subdivided into familial hemiplegic migraine (FHM) and sporadic hemiplegic migraine (SHM), distinguished by having a positive or negative family history for HM, respectively [1].
A subset of HM patients exhibits an autosomal dominant phenotype with single high-penetrant causal mutations present in ion transport genes CACNA1A, ATP1A2 or SCN1A [5,6,7]. However, in many HM patients, no such pathogenic mutation has been detected [8, 9]. Whereas evidence is accumulating that loss-of-function mutations in PRRT2 [10], a key component of the Ca2+-dependent neurotransmitter release machinery [11], are involved in HM, the gene more likely acts as a modifier of disease [12]. This suggests that HM, in a set of patients, may be regarded a complex disorder with multiple genetic factors contributing to the phenotype. Most relevant, a Finnish polygenic risk score study of genome-wide association study (GWAS) data has shown that HM patients without a high-penetrant disease-causing mutation in a known HM gene carry an excess of common (frequency > 1%) variants compared to patients suffering from common (complex) migraine subtypes [13].
Following along this line of evidence, it has been hypothesised that complex disorders can be the result of an accumulation of genetic variants in a disease pathway, where the crossing of a certain threshold leads to disease [14]. Moreover, current evidence indicates that complex traits are likely to be underpinned by a combination of multiple common and rare variants [15,16,17]. Here we set out to investigate the contribution of modulatory genetic effects that can be studied through testing the synergistic burden of (functional) variants, best annotated as missense variants, rather than a single causative mutation. Burden can be regarded as an accumulation of variants that are more often present in cases compared to controls. We hypothesise here that the burden of missense variants in certain ion channel genes might be involved in the disease pathology of HM.
CACNA1A was the first HM gene discovered and encodes the pore‐forming α1A subunit of the neuronal voltage‐gated calcium channel (VGCC) CaV2.1 (P/Q‐type) [5, 18]. CaV2.1 channels are predominantly localised at presynaptic terminals and play a prominent role in controlling neurotransmitter release at most synapses of the nervous system [19,20,21]. CACNA1A is a member of a family of rather conserved α1 subunit genes, hereafter referred to as “CACNA1x”, which are part of VGCCs that are classified as either high-voltage-activated (HVA) or low-voltage-activated (LVA) channels that are present on the membranes of excitable cells (Fig. 1) [22, 23]. CaV channels are typically composed of multiple subunits, namely an α1, a β, an α2/δ and a γ subunit. An α1 subunit has 24 transmembrane segments and forms the pore through which calcium ions pass into the cell. The main characteristic of the various CaV channel types is primarily determined by the type of α1 subunit, so the presence of either α1A, α1B, α1C, α1D, α1E, α1F, α1G, α1H, α1I or α1S. Given the important functions of CaV channels, it is not surprising that genetic variation in CACNA1x genes is not well-tolerated; the residual variation intolerance scores for these genes are high (Table 1) [24].
The voltage-gated calcium channel (VGCC) family of proteins. The α1 subunits can be divided into three subclasses according to their amino acid sequence identity, as shown in the dendrogram. CaV1 and CaV2 channels are high-voltage-activated (HVA), whereas CaV3 channels are low-voltage-activated (LVA). The genes encoding the respective α1 subunit are provided as well as the type of current the respective channel type produces. The schematic is based upon Perez-Reyes and Dolphin [22, 23]
The expression of CACNA1x genes varies considerably and, with the exception of CACNA1S, all are expressed in the brain [25]. In addition to CACNA1A being a well-known HM gene, there have been rare reports on other CACNA1x genes possibly involved in HM-relevant phenotypes. For instance, a link between hemiplegic migraine and brain stem aura migraine has been suggested for CACNA1E [26], and headache with neurological deficits and cerebrospinal fluid lymphocytosis (HaNDL), a headache syndrome with much phenotypic resemblance to HM, has been linked to the occurrence of antibodies against CACNA1H [27], a gene implicated in childhood epilepsy although this has recently been debated [28]. Furthermore, using a systems genetics approach, Rasmussen et al. [29] identified CACNA1B as one of the genes commonly mutated in migraine families. Finally, CACNA1A was identified as a risk locus for common migraine, as well as being one of the three genes specific for migraine with aura [30]. Although no definite proof for a causal link was provided in any of these cases, the existing data can be regarded as supportive evidence for a spectrum ranging from rare to common variants contributing to certain extent to the risk for both common and hemiplegic migraine. This variety of observed variants makes the family of CACNA1x genes an interesting candidate for burden testing in HM, with relevance, foremost, to patients with a complex genetic basis.
Whole exome sequencing (WES) enables comprehensive exploration of missense variants and investigation of their role in complex traits. When considering that these missense variants are unlikely to be causing HM as monogenic factors as occurs for patients with specific CACNA1A, ATP1A2 and SCN1A mutations [5,6,7], burden testing is a potential way to explore their potential synergistic effect on increasing HM disease risk. Burden testing typically requires a set of qualifying variants, often rare, protein-altering variants in the case of a monogenic condition. However, following on from the hypothesis that HM seems not a monogenic disorder in all patients, the accumulation of both rare and common protein-altering variants may be relevant in terms of disease susceptibility. The use of large publicly available WES datasets from general population controls can be incorporated in burden testing to gain more reliable estimates of gene-wide susceptibility.
We hypothesise that the burden of multiple missense variants in CACNA1x genes increases the risk for HM, burden being the aggregation of both rare and common variants as well as the increased presence of a variant in cases compared to controls. To this end, we here used WES data from a large Australian HM patient cohort to identify missense variants in eight calcium channel genes (CACNA1A, -B, -C, -D, -E, -G, -H and -I) and determined whether the aggregated effect of the variants across the genes was higher than observed in general population controls. Results were validated in an independent Dutch clinical HM cohort.
Methods
Study Cohorts
The study consisted of two cohorts of HM patients: an Australian cohort of 184 patients (discovery cohort) and a Dutch cohort of 32 patients (replication cohort). Importantly, patients were a priori excluded in case a pathogenic mutation was present in one of the three HM genes (CACNA1A, ATP1A2 and SCN1A) or HM-related genes with mutations confirmed by Sanger sequencing [9, 31].
Australian Cohort
The Australian cohort was selected out of over 300 patients that had been referred to the Genomics Research Centre (GRC) Diagnostic Clinic for genetic diagnostic testing after a suspected diagnosis of HM from the referring neurologist. From this cohort, a subset of 184 (122 females and 62 males) unrelated individuals tested negative for known HM gene mutations (CACNA1A, ATP1A2 and SCN1A) and HM-related genes [9, 31]. All cases consented to genetic testing with their doctors, as required under current regulations. Positive family history was reported for 25% of the cases; 5% were reported as SHM, while family information was not available for the remainder of cases.
DNA was extracted from blood samples using QIAGEN QIAamp DNA Mini Kit as per the manufacturers’ instructions. Next generation sequencing (NGS) libraries for WES were constructed using the Ion AmpliSeq™ Exome RDY library kits (ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. The Ion Chef was used to load sample libraries (barcoded fragments of 200 bp). WES was performed in the Genomics Research Centre (GRC), Australia via the Ion Proton and GeneStudio S5 plus (ThermoFisher Scientific) instruments using default settings for Ion AmpliSeq Exome RDY Kit 4 × 2 (Thermo Fisher Scientific). The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Human Research Ethics Committee of the Queensland University of Technology (approval number: 1800000611).
Dutch Cohort
The cohort consisted of 32 patients (22 females and 10 males) with FHM/SHM according to ICHD-3 criteria [32]. Patients were selected from the Leiden Headache Centre at the Leiden University Medical Centre (LUMC), and contained patients (i) seen in person by experienced headache clinicians or research physicians or (ii) referred from elsewhere for clinical genetic research with records being evaluated and clinical diagnosis confirmed by GMT, NP and IdB [4]. All patients were from different families and did not have a known pathogenic mutation in one of the three HM genes. The study was approved by the Medical Ethics Committee of LUMC and all participants provided informed consent.
Genomic DNA was extracted from peripheral blood leukocytes according to the standard salting-out protocol [33]. WES was performed using in-house sequencing facility (Leiden Genome Technology Centre; URL: lgtc.nl) or outsourced to the Beijing Genomics Institute sequencing facility (URL: bgi.com). In brief, for the LGCT, coding sequences in the DNA were enriched using the SureSelect Human All Exon 50 Mb kit (Agilent Technologies, Santa Clara, CA, USA). Following sequence capture and amplification, fragments were sequenced using the Illumina HiSeq2000 platform (San Diego, CA, USA).
Controls
As a control dataset, we used summary statistics from gnomAD (see below in the paragraph on TRAPD methods). The gnomAD database was chosen as it consists of a large number of individuals and contains a detailed catalogue of exome-wide genetic variation. Furthermore, gnomAD provided ancestry information. The gnomAD database contains exome variant summary statistics for 56,885 non-Finnish Europeans, with a female-to-male ratio of ~ 1.27:1, depending on the available genotypes at each specific locus. As HM is a very rare disorder with a prevalence of 0.01% [34], confounding effects due to the presence of HM patients in the control group were deemed to be negligible.
Whole Exome Sequencing and Quality Control
Australian Cohort
Following WES, the Ion Torrent Server was used to generate quality metrics, align reads to the Human Genome 19 (Hg19), and the Ion Torrent Variant Caller (TVC) was used to call sequence variants and produce variant calling format (VCF) files.
Dutch Cohort
Following sequencing, the sequence reads were aligned to the UCSC Genome Browser hg19 reference sequence using the Burrows-Wheeler Alignment tool [35]. The generated BAM files were subsequently converted to VCF files using BCFtools [36].
Single-Variant Analysis
Prior to performing burden testing, all variants were assessed to determine whether there were obvious, high-penetrant disease-causing mutations detected outside of the known HM genes that could cause HM in patients of either cohort. In the absence of such pathogenic mutation, individual missense variants in all the CACNA1x genes were assessed for patients of the Australian cohort. For the Dutch cohort, only those missense variants present in TRAPD-associated CACNA1x genes were investigated.
Burden Testing
Variants Pre-processing All Cohorts
As VCFs were exported from different platforms, the respective analyses had to be unified. The first step for both cohorts was to normalise VCFs using BCFtool; this ensures that any platform-specific formatting differences are removed and also expands multi-allelic variants [36]. VCFs were merged for each cohort using vcftools, and variants with average read depth coverage below × 10 were excluded using either BCFtools or the snpEff program [36, 37]. For both cohorts, the coding exons of the CACNA1x genes were included with a 5-bp pad on either side of the exon. New VCFs (one merged for each cohort) were annotated with VEP Ensembl [38]. For the Dutch cohort as an extra quality control step, only those variants with a quality-by-depth (QD) score > 4 were taken forward.
Selection of Qualifying Variants
To determine the number of variants, we selected “qualifying variants” being variants that meet the criteria of inclusion. Only those variants classified (annotated) as missense variants were considered as “qualifying variants”. The number of individuals in the case cohort who carried at least one “qualifying variant” in that gene and the total number of variants were used in the analysis. For the gnomAD control dataset, only summary statistics were available. Therefore, to approximate the number of control subjects carrying at least one qualifying variant in a given gene, the allele counts for all qualifying variants in that gene were summed. This summation-based approximation probably is an overestimation as it is likely that some individuals carry multiple variants in the same gene. Contrary to rare variant analysis where only the locations of the qualifying variant in cases are used for controls, we selected all variants across the entire gene in controls, in the same way as what was done for the cases. As a result, we had a total number of all missense variants per CACNA1x gene in both cases and controls. These “qualifying variants” for both the case and the control cohort were compared. Insertions and deletions (Indels) were not included in the analysis due to their higher percentage of sequencing artefacts, especially given the differing sequencing platforms used across cohorts.
Multiple-Variant Burden Testing of CACNA1x Genes
Gene-based burden testing was performed for all variants that met the quality filters, which are referred to as “qualifying variants”, using adaptation on the TRAPD test (Testing Rare vAriants using Public Data) [39]. TRAPD was chosen because the control dataset consisted of summary data rather than individual-level genotype data as well as for its approach to collate variants which mitigates the statistical drawbacks of burden testing per variant or per individual. The TRAPD test was implemented to determine whether CACNA1x genes and subjects carried a significant burden of missense variants in cases. TRAPD produces counts of “collapsed” variant groups across each gene and for the respective case or control cohort. To conduct the test, a group file with the qualifying variants was created for each of the eight genes (CACNA1A, CACNA1B, CACNA1C, CACNA1D, CACNA1E, CACNA1G, CACNA1H and CACNA1I). Of note, CACNA1S was excluded from the analysis as it encodes the pore-forming CaV1.1 α1S subunit that is exclusively expressed in skeletal muscle, so not in the brain and CACNA1F was excluded as this gene is located on the X-chromosome, and TRAPD is currently not configured to test non-autosomal chromosomes.
We performed gene-based burden testing for all single-point variants in each cohort. The following steps, in brief, were performed: (1) variants for each CACNA1x gene in the case group were compiled into a “SNP file”, (2) a Python script was used to interrogate the VCFs and count the occurrence of variants in each gene in both the case and the control cohorts independently. This generated variant count data for each gene, and (3) the one-sided Fisher exact test was used on the allele count tables to identify the probability of excess in the number of allele counts in cases relative to controls (i.e. the statistical significance of the burden). (4) The one-sided Fisher exact test was used on the subject count tables to identify the probability of excess in the number of subjects with variants in cases relative to controls (i.e., the statistical significance of the burden). P-values < 6.25 × 10−3 were considered significant (Bonferroni corrected for testing 8 genes). Odd ratios were calculated to assess the magnitude of the burden effect. Genes exhibiting statistically significant burden in HM from the Australian discovery cohort were also tested in the Dutch replication cohort.
Results
Single-Variant Analysis
No clear pathogenic mutations in CACNA1x genes were identified from the WES data in patients from either the Australian or the Dutch cohorts. However, the number of variants in CACNA1x genes prompted us to perform burden testing. In the Australian cohort, we identified 79 different missense variants in the eight CACNA1x genes examined in the 184 HM patient group from Australia (Supplementary Table 1). All but seven of the variants had been previously identified (i.e., they have an rs number in dbSNP). The seven novel variants were all single-case across multiple different CACNA1x genes. In the Dutch cohort, four different variants were identified in CACNA1I and ten in CACNA1H; all of which had been previously identified (Supplementary Table 2). Although some missense variants in CACNA1x genes were predicted to have a pathogenic potential, there was not enough evidence for causality in a monogenic manner such as has been shown for the three well-known HM genes. The results of the individual variant analyses indicate the existence of many variants across CACNA1x genes that in combination could plausibly confer increased susceptibility to HM, especially when considered collectively using burden analysis.
Multiple-Variant Burden Testing of CACNA1x Genes
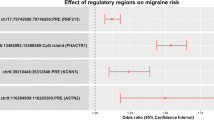
Considering the results from the individual variant analysis, a multiple-variant burden analysis was performed to test whether there is an over-representation of CACNA1x missense variants in HM compared to controls. As shown in Table 2, in the Australian HM cohort, this analysis revealed a significantly increased burden of missense variants for those with HM in CACNA1E (p = 1.3 × 10−4), CACNA1H (p < 2.2 × 10−16) and CACNA1I (p < 2.2 × 10−16). For the Dutch cohort, this replicated for CACNA1H (p = 3.5 × 10−8, pcor = 1.04 × 10−7) and CACNA1I (p = 0.019, pcor = 0.056), but not CACNA1E (p = 0.85), albeit that CACNA1I did not remain significant after correction for multiple testing. In addition, the number of subjects carrying a variant was also higher cases in CACNA1E (p = 6.2 × 10−3), CACNA1H (p < 2.2 × 10−16) and CACNA1I (p < 2.2 × 10−16) (Table 2). The results showed evidence of replication in the Dutch cohort for CACNA1H (p = 1.2 × 10−2; pcor = 3.6 × 10−2) and CACNA1I (p = 4.4 × 10−2; pcor = 0.13), but not for CACNA1E (p = 0.88), albeit CACNA1I did not remain significant after correction for multiple testing. All but four variants were outside the transmembrane domains that are typically affected in case of a pathogenic mutation in either CaV channel (Fig. 2).
Schematic representation of the α1 subunit, with the position of identified variants in this study, of the CaV3.2 and the CaV3.3 channels, encoded by the CACNA1H (A) and CACNA1I (B), respectively. Variants identified in the Australian HM cohort are depicted with a green dot, and variants identified in the Dutch HM cohort are depicted with a red dot
Discussion
Here we used WES data from 184 suspected HM patients from an Australian clinically referred cohort and compared these to the publicly available gnomAD control dataset using TRAPD, finding that CACNA1E, CACNA1I and CACNA1H missense variants were more prevalent in cases. Furthermore, we show evidence for replication of these findings for CACNA1H and CACNA1I in a Dutch clinical HM cohort. This finding emphasises that although the cohorts differ in terms of inclusion criteria, the results are transferable to both groups.
In the general population, females are overrepresented in most forms of migraine including hemiplegic migraine. The overall female to male sex ratio in our HM cohort was ~ 1.97:1. This observed difference in prevalence will in part be explained by the fact that females are more inclined to consult a physician and thus are diagnosed earlier and more often than males [40]. We cannot rule out that there is any sexual dimorphic effect at any of the CACNA1x genes (i.e., a sex bias in gene function), but we consider this a minor factor compared to the ascertainment bias.
We have hypothesised that HM may not be autosomal dominant in a substantial proportion of cases, but rather is genetically a more complex trait. The difficulty in confirming this hypothesis lies in how to identify such variants, as they are neither identified by gene association approaches, nor in genome-wide association studies (GWAS). In order to identify such variants, we have used a methodology adapted from a TRAPD analysis. The methodology has proven itself by the identification of functional genetic variants in idiopathic hypogonadotropic hypogonadism [39]. By slightly adapting the method, we have been able to investigate all missense variance and thereby determine the variant and subject burden. Similarly, our results show that the accumulation of missense variants in CACNA1H and CACNA1I plays a role in HM.
CACNA1H and CACNA1I encode the α1 subunits of CaV3.2 and CaV3.3 LVA T-type calcium channels, respectively (Fig. 1) [22, 23]. CACNA1H is expressed ubiquitously, whereas CACNA1I is predominantly expressed in the brain [25]. CaV3.2 and CaV3.3 LVA T-type calcium channels [41], which open by only a small membrane depolarization, coupled with their tonic inactivation near resting membrane potential, underlie the spike/rebound bursting phenomenon seen with many types of neurons [42, 43]. These channels are localised at presynaptic nerve terminals [44] where they control synaptic transmission by directly triggering the release of neurotransmitters [45,46,47]. Inactivation of Cacna1h in mice led to decreased nociceptive signalling [48, 49] and several neurological symptoms [50, 51], whereas Cacna1i knock-out mice, and also Cacna1i/Cacna1h double knockout mutants, show implications for sleep rhythmogenesis [52]. T-type channels are important for human physiology, so mutations in these channels may lead, at least in theory, to channelopathies with clinical manifestations resulting from aberrant biophysical characteristics and/or cell surface trafficking issues of channels due to a gain or loss of channel function. Indeed, specific missense variants in CACNA1H have been implicated in a range of human conditions [50], including autism spectrum disorders [53] and amyotrophic lateral sclerosis [54]. Many missense variants in the human CACNA1H gene have been reported in patients presenting with a range of epilepsy syndromes [50], so the gene was labelled a risk gene for idiopathic generalised epilepsies (38). Functional analyses in embryonic kidney cells, however, revealed that the variants in CACNA1H generally produce mild biophysical changes and in some cases do not alter the gating of the channel and variants do not segregate with the phenotype [50]. Hence, their contribution to human epilepsies should be debated, as was recently suggested [28]. In line with this suggestion, it is not unexpected that CACNA1H variants identified in HM patients also not solely cause disease, although a burden of variants in this gene can still contribute to HM risk. Similarly, CACNA1I loss-of-function variants were identified that alter the gating properties of CaV3.3 channels, disrupt neuronal excitability and network activity, and have been associated with risk of developing schizophrenia and a range of neurodevelopmental disorders featuring developmental delay and epilepsy [55, 56]. Moreover, using patch-clamp electrophysiology, we have shown various functional alterations of channel activity for selected Cav3.3 rare variants, providing further evidence that CACNA1I may play a role in the development of HM [57].
Hence, the most likely scenario is that an increased burden of missense variants in CACNA1H and CACNA1I acts as a genetic modifier of disease risk. Such a modification of risk is not different when reviewing mutations that have been identified in some HM patients in a number of genes, including PRRT2 [12], PNKD [58], SLC4A4 [59], SLC1A3 [60] and SLC2A1 [61], that are primarily associated with movement or solute transport disorders.
Our study has some limitations. First of all, contrary to what is commonly undertaken in genetics, we considered both rare and common variants as an overarching burden of missense variants in this study. To support the validity of this approach, we used the Dutch replication cohort to validate findings from the Australian cohort. Further replication efforts in other independent cohorts would be of benefit in future studies of these genes. Secondly, as this study is the first of its kind, we narrowed the genes targeted to CACNA1x ion channels, due to known association of genes of this family with HM. However, the burden of variants in additional genes is likely to play a role in determining HM disease risk. Thirdly, we have used summary statistics for the controls that prevented us to compare ancestry of cases and controls together, although we ensured that both cases and controls were of European ancestry. Finally, the use of the gnomAD population as a control cohort means that we are not comparing truly matched populations. Both in our case cohorts and the gnomAD cohort, there are slightly more female than male participants, that is, the female to male ratio for the cases is ~ 1.97:1, and for the controls, it is ~ 1.27:1, which may result in a slight confounding effect, as does differences in environmental and cultural differences that could not be controlled in our study.
Conclusion
This study provides evidence that increased burden of missense variants in the amount of variants and the number of subjects carrying a variant in CACNA1H and CACNA1I exists for HM, and that these genes can modify HM disease risk, supporting more complex types of heritability for HM, in addition to the strictly monogenic forms.
Data Availability
The data used and/or analysed during the current study are available from the corresponding author on reasonable request.
Change history
23 February 2023
Missing Open Access funding information has been added in the Funding Note.
References
(2018) Headache Classification Committee of the International Headache Society (IHS) The international classification of headache disorders, 3rd edition. Cephalalgia 38:1–211. https://doi.org/10.1177/0333102417738202
Ducros A, Denier C, Joutel A, Cecillon M, Lescoat C, Vahedi K, Darcel F, Vicaut E et al (2001) The clinical spectrum of familial hemiplegic migraine associated with mutations in a neuronal calcium channel. N Engl J Med 345:17–24. https://doi.org/10.1056/NEJM200107053450103
Russell MB, Ducros A (2011) Sporadic and familial hemiplegic migraine: pathophysiological mechanisms, clinical characteristics, diagnosis, and management. Lancet Neurol 10:457–470. https://doi.org/10.1016/S1474-4422(11)70048-5
Pelzer N, Haan J, Stam AH, Vijfhuizen LS, Koelewijn SC, Smagge A, de Vries B, Ferrari MD et al (2018) Clinical spectrum of hemiplegic migraine and chances of finding a pathogenic mutation. Neurology 90:e575–e582. https://doi.org/10.1212/WNL.0000000000004966
Ophoff RA, Terwindt GM, Vergouwe MN, van Eijk R, Oefner PJ, Hoffman SM, Lamerdin JE, Mohrenweiser HW et al (1996) Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell 87:543–552. https://doi.org/10.1016/s0092-8674(00)81373-2
Dichgans M, Freilinger T, Eckstein G, Babini E, Lorenz-Depiereux B, Biskup S, Ferrari MD, Herzog J et al (2005) Mutation in the neuronal voltage-gated sodium channel SCN1A in familial hemiplegic migraine. Lancet 366:371–377. https://doi.org/10.1016/S0140-6736(05)66786-4
De Fusco M, Marconi R, Silvestri L, Atorino L, Rampoldi L, Morgante L, Ballabio A, Aridon P et al (2003) Haploinsufficiency of ATP1A2 encoding the Na+/K+ pump alpha2 subunit associated with familial hemiplegic migraine type 2. Nat Genet 33:192–196. https://doi.org/10.1038/ng1081
Hiekkala ME, Vuola P, Artto V, Happola P, Happola E, Vepsalainen S, Cuenca-Leon E, Lal D et al (2018) The contribution of CACNA1A, ATP1A2 and SCN1A mutations in hemiplegic migraine: a clinical and genetic study in Finnish migraine families. Cephalalgia 38:1849–1863. https://doi.org/10.1177/0333102418761041
Sutherland HG, Maksemous N, Albury CL, Ibrahim O, Smith RA, Lea RA, Haupt LM, Jenkins B et al (2020) Comprehensive exonic sequencing of hemiplegic migraine-related genes in a cohort of suspected probands identifies known and potential pathogenic variants. Cells 9. https://doi.org/10.3390/cells9112368
Riant F, Roos C, Roubertie A, Barbance C, Hadjadj J, Auvin S, Baille G, Beltramone M et al (2022) Hemiplegic migraine associated with PRRT2 variations: a clinical and genetic study. Neurology 98:e51–e61. https://doi.org/10.1212/WNL.0000000000012947
Valente P, Castroflorio E, Rossi P, Fadda M, Sterlini B, Cervigni RI, Prestigio C, Giovedi S et al (2016) PRRT2 is a key component of the Ca(2+)-dependent neurotransmitter release machinery. Cell Rep 15:117–131. https://doi.org/10.1016/j.celrep.2016.03.005
Pelzer N, de Vries B, Kamphorst JT, Vijfhuizen LS, Ferrari MD, Haan J, van den Maagdenberg AM, Terwindt GM (2014) PRRT2 and hemiplegic migraine: a complex association. Neurology 83:288–290. https://doi.org/10.1212/WNL.0000000000000590
Gormley P, Kurki M, Kurki MI, Hiekkala M, Hiekkala ME, Veerapen K, Häppölä P, Mitchell AA et al (2018) Common variant burden contributes to the familial aggregation of migraine in 1,589 families. Neuron 98:743-753.e4. https://doi.org/10.1016/j.neuron.2018.04.014
Wray NR, Goddard ME (2010) Multi-locus models of genetic risk of disease. Genome Med 2:10. https://doi.org/10.1186/gm131
Gudmundsson J, Sulem P, Gudbjartsson DF, Masson G, Agnarsson BA, Benediktsdottir KR, Sigurdsson A, Magnusson OT et al (2012) A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat Genet 44:1326–1329. https://doi.org/10.1038/ng.2437
Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S et al (2011) Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet 43:1066–1073. https://doi.org/10.1038/ng.952
Panoutsopoulou K, Tachmazidou I, Zeggini E (2013) In search of low-frequency and rare variants affecting complex traits. Hum Mol Genet 22:R16-21. https://doi.org/10.1093/hmg/ddt376
Ferrari MDP, Klever RRM, Terwindt GMMD, Ayata CMD, van den Maagdenberg AMJMP (2015) Migraine pathophysiology: lessons from mouse models and human genetics. Lancet Neurol 14:65–80. https://doi.org/10.1016/S1474-4422(14)70220-0
Westenbroek RE, Sakurai T, Elliott EM, Hell JW, Starr TV, Snutch TP, Catterall WA (1995) Immunochemical identification and subcellular distribution of the alpha 1A subunits of brain calcium channels. J Neurosci 15:6403–6418
Catterall WA (1998) Structure and function of neuronal Ca2+ channels and their role in neurotransmitter release. Cell Calcium 24:307–323. https://doi.org/10.1016/s0143-4160(98)90055-0
Dunlap K, Luebke JI, Turner TJ (1995) Exocytotic Ca2+ channels in mammalian central neurons. Trends Neurosci 18:89–98
Perez-Reyes E (2003) Molecular physiology of low-voltage-activated t-type calcium channels. Physiol Rev 83:117–161. https://doi.org/10.1152/physrev.00018.2002
Dolphin AC (2012) Calcium channel auxiliary α2δ and β subunits: trafficking and one step beyond. Nat Rev Neurosci 13:542–555. https://doi.org/10.1038/nrn3311
Petrovski S, Gussow AB, Wang Q, Halvorsen M, Han Y, Weir WH, Allen AS, Goldstein DB (2015) The intolerance of regulatory sequence to genetic variation predicts gene dosage sensitivity. PLoS Genet 11:e1005492. https://doi.org/10.1371/journal.pgen.1005492
(2013) The Genotype-Tissue Expression (GTEx) project. Nat Genet 45:580–5. https://doi.org/10.1038/ng.2653
Ambrosini A, D’Onofrio M, Buzzi MG, Arisi I, Grieco GS, Pierelli F, Santorelli FM, Schoenen J (2017) Possible involvement of the CACNA1E gene in migraine: a search for single nucleotide polymorphism in different clinical phenotypes. Headache 57:1136–1144. https://doi.org/10.1111/head.13107
Kurtuncu M, Kaya D, Zuliani L, Erdag E, Icoz S, Ugurel E, Cavus F, Aysit N et al (2013) CACNA1H antibodies associated with headache with neurological deficits and cerebrospinal fluid lymphocytosis (HaNDL). Cephalalgia Int J Headache 33:123–129. https://doi.org/10.1177/0333102412463494
Calhoun JD, Huffman AM, Bellinski I, Kinsley L, Bachman E, Gerard E, Kearney JA, Carvill GL (2020) CACNA1H variants are not a cause of monogenic epilepsy. Hum Mutat 41:1138–1144. https://doi.org/10.1002/humu.24017
Rasmussen AH, Kogelman LJA, Kristensen DM, Chalmer MA, Olesen J, Hansen TF (2020) Functional gene networks reveal distinct mechanisms segregating in migraine families. Brain J Neurol 143:2945–2956. https://doi.org/10.1093/brain/awaa242
Hautakangas H, Winsvold BS, Ruotsalainen SE, Bjornsdottir G, Harder AVE, Kogelman LJA, Thomas LF, Noordam R et al (2022) Genome-wide analysis of 102,084 migraine cases identifies 123 risk loci and subtype-specific risk alleles. Nat Genet. https://doi.org/10.1038/s41588-021-00990-0
Maksemous N, Smith RA, Sutherland HG, Maher BH, Ibrahim O, Nicholson GA, Carpenter EP, Lea RA et al (2019) Targeted next generation sequencing identifies a genetic spectrum of DNA variants in patients with hemiplegic migraine. Cephalalgia Rep 2:2515816319881630. https://doi.org/10.1177/2515816319881630
(2013) The international classification of headache disorders, 3rd edition (beta version). Cephalalgia 33:629–808. https://doi.org/10.1177/0333102413485658
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215. https://doi.org/10.1093/nar/16.3.1215
Thomsen LL, Eriksen MK, Roemer SF, Andersen I, Olesen J, Russell MB (2002) A population-based study of familial hemiplegic migraine suggests revised diagnostic criteria. Brain 125:1379–1391. https://doi.org/10.1093/brain/awf132
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T et al (2021) Twelve years of SAMtools and BCFtools. Gigascience 10. https://doi.org/10.1093/gigascience/giab008
Cingolani P, Platts A, le Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu X et al (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. https://doi.org/10.4161/fly.19695
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F (2016) The Ensembl variant effect predictor. Genome Biol 17:122. https://doi.org/10.1186/s13059-016-0974-4
Guo MH, Plummer L, Chan YM, Hirschhorn JN, Lippincott MF (2018) Burden testing of rare variants identified through exome sequencing via publicly available control data. Am J Hum Genet 103:522–534. https://doi.org/10.1016/j.ajhg.2018.08.016
Rasmussen BK, Jensen R, Olesen J (1992) Impact of headache on sickness absence and utilisation of medical services: a Danish population study. J Epidemiol Community Health 46:443–446. https://doi.org/10.1136/jech.46.4.443
Ertel EA, Campbell KP, Harpold MM, Hofmann F, Mori Y, Perez-Reyes E, Schwartz A, Snutch TP et al (2000) Nomenclature of voltage-gated calcium channels. Neuron 25:533–535. https://doi.org/10.1016/s0896-6273(00)81057-0
Chemin J, Monteil A, Perez-Reyes E, Bourinet E, Nargeot J, Lory P (2002) Specific contribution of human T-type calcium channel isotypes (alpha(1G), alpha(1H) and alpha(1I)) to neuronal excitability. J Physiol 540:3–14. https://doi.org/10.1113/jphysiol.2001.013269
Cain SM, Snutch TP (2010) Contributions of T-type calcium channel isoforms to neuronal firing. Channels (Austin Tex) 4:475–482. https://doi.org/10.4161/chan.4.6.14106
Huang Z, Lujan R, Kadurin I, Uebele VN, Renger JJ, Dolphin AC, Shah MM (2011) Presynaptic HCN1 channels regulate Cav3.2 activity and neurotransmission at select cortical synapses. Nat Neurosci 14:478–486. https://doi.org/10.1038/nn.2757
Pan ZH, Hu HJ, Perring P, Andrade R (2001) T-type Ca(2+) channels mediate neurotransmitter release in retinal bipolar cells. Neuron 32:89–98. https://doi.org/10.1016/s0896-6273(01)00454-8
Weiss N, Zamponi GW (2013) Control of low-threshold exocytosis by T-type calcium channels. Biochim Biophys Acta 1828:1579–1586. https://doi.org/10.1016/j.bbamem.2012.07.031
Zamponi GW, Striessnig J, Koschak A, Dolphin AC (2015) The physiology, pathology, and pharmacology of voltage-gated calcium channels and their future therapeutic potential. Pharmacol Rev 67:821–870. https://doi.org/10.1124/pr.114.009654
Choi S, Na HS, Kim J, Lee J, Lee S, Kim D, Park J, Chen CC et al (2007) Attenuated pain responses in mice lacking Ca(V)3.2 T-type channels. Genes Brain Behav 6:425–431. https://doi.org/10.1111/j.1601-183X.2006.00268.x
Bourinet E, Alloui A, Monteil A, Barrere C, Couette B, Poirot O, Pages A, McRory J et al (2005) Silencing of the Cav3.2 T-type calcium channel gene in sensory neurons demonstrates its major role in nociception. EMBO J 24:315–324. https://doi.org/10.1038/sj.emboj.7600515
Weiss N, Zamponi GW (2020) Genetic T-type calcium channelopathies. J Med Genet 57:1–10. https://doi.org/10.1136/jmedgenet-2019-106163
Gangarossa G, Laffray S, Bourinet E, Valjent E (2014) T-type calcium channel Cav3.2 deficient mice show elevated anxiety, impaired memory and reduced sensitivity to psychostimulants. 8. https://doi.org/10.3389/fnbeh.2014.00092
Astori S, Wimmer RD, Prosser HM, Corti C, Corsi M, Liaudet N, Volterra A, Franken P et al (2011) The Ca(V)3.3 calcium channel is the major sleep spindle pacemaker in thalamus. Proc Natl Acad Sci USA 108:13823–13828. https://doi.org/10.1073/pnas.1105115108
Splawski I, Yoo DS, Stotz SC, Cherry A, Clapham DE, Keating MT (2006) CACNA1H Mutations in autism spectrum disorders*. J Biol Chem 281:22085–22091. https://doi.org/10.1074/jbc.M603316200
Carter MT, McMillan HJ, Tomin A, Weiss N (2019) Compound heterozygous CACNA1H mutations associated with severe congenital amyotrophy. Channels (Austin) 13:153–161. https://doi.org/10.1080/19336950.2019.1614415
Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H et al (2013) Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154:518–529. https://doi.org/10.1016/j.cell.2013.06.049
El Ghaleb Y, Schneeberger PE, Fernandez-Quintero ML, Geisler SM, Pelizzari S, Polstra AM, van Hagen JM, Denecke J et al (2021) CACNA1I gain-of-function mutations differentially affect channel gating and cause neurodevelopmental disorders. Brain 144:2092–2106. https://doi.org/10.1093/brain/awab101
Maksemous N, Blayney CD, Sutherland HG, Smith RA, Lea RA, Tran KN, Ibrahim O, McArthur JR et al (2022) Investigation of CACNA1I Cav3.3 dysfunction in hemiplegic migraine. Front Mol Neurosci 15:892820. https://doi.org/10.3389/fnmol.2022.892820
Gardiner AR, Jaffer F, Dale RC, Labrum R, Erro R, Meyer E, Xiromerisiou G, Stamelou M et al (2015) The clinical and genetic heterogeneity of paroxysmal dyskinesias. Brain 138:3567–3580. https://doi.org/10.1093/brain/awv310
Suzuki M, Van Paesschen W, Stalmans I, Horita S, Yamada H, Bergmans BA, Legius E, Riant F et al (2010) Defective membrane expression of the Na(+)-HCO(3)(-) cotransporter NBCe1 is associated with familial migraine. Proc Natl Acad Sci USA 107:15963–15968. https://doi.org/10.1073/pnas.1008705107
Jen JC, Wan J, Palos TP, Howard BD, Baloh RW (2005) Mutation in the glutamate transporter EAAT1 causes episodic ataxia, hemiplegia, and seizures. Neurology 65:529–534. https://doi.org/10.1212/01.wnl.0000172638.58172.5a
Weller CM, Leen WG, Neville BG, Duncan JS, de Vries B, Geilenkirchen MA, Haan J, Kamsteeg EJ et al (2015) A novel SLC2A1 mutation linking hemiplegic migraine with alternating hemiplegia of childhood. Cephalalgia 35:10–15. https://doi.org/10.1177/0333102414532379
Acknowledgements
We greatly acknowledge the subjects who participated in this study.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. The authors disclose receipt of the following financial support for the research, authorship, and/or publication of this article. This work was supported by the Australian National Health and Medical Research Council (NHMRC-APP1122387) (LRG); a Migraine Research Foundation grant (2016), NY, USA (LRG); an Australian International Science Linkages grant (LRG); by infrastructure purchased with Australian Government EIF Super Science Funds as part of the Therapeutic Innovation Australia – Queensland Node project (LRG); the Centre of Medical System Biology (CMSB) in the framework of the Netherlands Genomics Initiative (NGI) 050–060-409 (AMJMvdM); and the European Community (EC) FP7-EUROHEADPAIN (no. 602633; AMJMvdM).
Author information
Authors and Affiliations
Contributions
Omar Ibrahim, Aster V. E. Harder, Lisanne S. Vijfhuizen, Rodney A. Lea, Arn M. J. M. van den Maagdenberg and Lyn R. Griffiths contributed to the study conception and design. Whole exome sequencing and data collection were performed by Neven Maksemous, Heidi Sutherland, Nadine Pelzer, Irene de Boer and Gisela M. Terwindt. Material preparation and analysis were performed by Omar Ibrahim, Aster V. E. Harder, Neven Maksemous, Lisanne S. Vijfhuizen and Rodney A. Lea. The first draft of the manuscript was written by Omar Ibrahim and Aster V. E. Harder with additional input from all co-authors. The final manuscript was read and approved by all authors.
Corresponding authors
Ethics declarations
Ethics Approval
The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Human Research Ethics Committee of the Queensland University of Technology (approval number: 1800000611) and by the Medical Ethics Committee of LUMC.
Consent to Participate
Appropriate consents for the patient cohort are already in place.
Consent for Publication
Not applicable.
Competing Interests
GMT reports consultancy support from Novartis, Allergan/Abbvie, Lilly, and Teva, Lundbeck and independent support from Dutch Research Council, the Dutch Heart & Brain Foundations, IRRF and Dioraphte. AMJMvdM reports research support from Praxis Precision Medicine and Schedule 1 Therapeutics and consultancy support from AbbVie. LRG reports recent consultancy support from Teva and research support from the Australian National Health and Medical Research Council and the US Migraine Research Foundation. IdB reports independent support from the International Retinal Research Foundation and Dutch Heart Foundation. AVEH, LSV, OI, NP, NM, HS and RAL declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Maksemous, N., Harder, A.V.E., Ibrahim, O. et al. Whole Exome Sequencing of Hemiplegic Migraine Patients Shows an Increased Burden of Missense Variants in CACNA1H and CACNA1I Genes. Mol Neurobiol 60, 3034–3043 (2023). https://doi.org/10.1007/s12035-023-03255-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-023-03255-5