Abstract
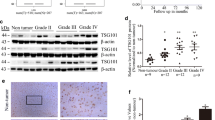
The invasion of glioblastoma usually results in the recurrence and poor prognosis in patients with glioma. However, the underlying mechanisms involved in glioma invasion remains undefined. In this study, immunohistochemistry analyses of glioma specimens demonstrated that high expression of Par6 was positively correlated with malignancy and poor prognosis of patients with glioma. Par6-overexpressing glioma cells showed much more fibroblast-like morphology, suggesting that regulation of Par6 expression might be associated with tumor invasion in glioma cells. Further study indicated that Par6 overexpression subsequently increased CD44 and N-cadherin expression to enhance glioma invasion through activating MEK/ERK/STAT3 pathway, in vivo and in vitro. Moreover, we found that LIN28/let-7d axis was involved in this process via a positive feedback loop, suggesting that MEK/ERK/LIN28/let-7d/STAT3 cascade might be essential for Par6-mediated glioma invasion. Therefore, these data highlight the roles of Par6 in glioma invasion, and Par6 may serve as a potential therapeutic target for patients with glioma.








Similar content being viewed by others
Data Availability
All routine analysis methods are included in the “Methods” section. Sequencing data that supporting the findings of this study have been deposited in the Gene Expression Omnibus database under accession code SUB6244767. The data that support the findings of this study are presented in the paper, and all raw data are available from the corresponding author upon reasonable request. The published miRbase dataset used in this study were obtained from the publicly available database.
Abbreviations
- CNS:
-
Central nervous system
- GBM:
-
Glioblastoma
- EMT:
-
Epithelial-to-mesenchymal transition
- ECM:
-
Extracellular matrix
- TJs:
-
Tight junctions
- TCGA:
-
The Cancer Genome Atlas
- ERK:
-
Extracellular signal-regulated kinase
- NSCLC:
-
Non-small-cell lung cancer
- STR:
-
Short tandem repeat
- IHC:
-
Immunohistochemistry
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- H&E:
-
Hematoxylin-eosin
- TGF-β:
-
Transforming growth factor-β
- TMA:
-
Tissue microarray
- MMP:
-
Matrix metalloproteinase
- DEGs:
-
Differentially expressed genes.
References
Shi J, Dong B, Cao J, Mao Y, Guan W, Peng Y et al (2017) Long non-coding RNA in glioma: signaling pathways. Oncotarget 8:27582–27592
Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS (2018) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Neuro-Oncology 20:iv1–iv86
Jansen M, Yip S, Louis DN (2010) Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol 9:717–726
Rock K, McArdle O, Forde P, Dunne M, Fitzpatrick D, O'Neill B et al (2012) A clinical review of treatment outcomes in glioblastoma multiforme--the validation in a non-trial population of the results of a randomised Phase III clinical trial: has a more radical approach improved survival? Br J Radiol 85:e729–e733
Thiery JP (2002) Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2:442–454
Thiery JP, Sleeman JP (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 7:131–142
Cox TR, Erler JT (2011) Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis Model Mech 4:165–178
Lu P, Weaver VM, Werb Z (2012) The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol 196:395–406
Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME (2007) Molecular targets of glioma invasion. Cell Mol Life Sci 64:458–478
Yuan Y, Li SL, Cao YL, Li JJ, Wang QP (2019) LKB1 suppresses glioma cell invasion via NF-kappaB/Snail signaling repression. Oncotargets Ther 12:2451–2463
Ou Y, Wu Q, Wu C, Liu X, Song Y, Zhan Q (2017) Migfilin promotes migration and invasion in glioma by driving EGFR and MMP-2 signalings: a positive feedback loop regulation. J Genet Genomics 44:557–565
Watts JL, Etemad-Moghadam B, Guo S, Boyd L, Draper BW, Mello CC et al (1996) par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development 122:3133–3140
Bose R, Wrana JL (2006) Regulation of Par6 by extracellular signals. Curr Opin Cell Biol 18:206–212
Etienne-Manneville S, Hall A (2001) Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell 106:489–498
Kim M, Datta A, Brakeman P, Yu W, Mostov KE (2007) Polarity proteins PAR6 and aPKC regulate cell death through GSK-3beta in 3D epithelial morphogenesis. J Cell Sci 120:2309–2317
Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E (2003) ACF7: an essential integrator of microtubule dynamics. Cell 115:343–354
Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME (2004) Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci 7:1195–1203
Aranda V, Nolan ME, Muthuswamy SK (2008) Par complex in cancer: a regulator of normal cell polarity joins the dark side. Oncogene 27:6878–6887
Marques E, Klefstrom J (2015) Par6 family proteins in cancer. Oncoscience 2:894–895
Nolan ME, Aranda V, Lee S, Lakshmi B, Basu S, Allred DC et al (2008) The polarity protein Par6 induces cell proliferation and is overexpressed in breast cancer. Cancer Res 68:8201–8209
Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL (2005) Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science 307:1603–1609
Viloria-Petit AM, David L, Jia JY, Erdemir T, Bane AL, Pinnaduwage D et al (2009) A role for the TGFbeta-Par6 polarity pathway in breast cancer progression. Proc Natl Acad Sci U S A 106:14028–14033
Avery-Cooper G, Doerr M, Gilbert RW, Youssef M, Richard A, Huether P et al (2014) Par6 is an essential mediator of apoptotic response to transforming growth factor beta in NMuMG immortalized mammary cells. Cancer Cell Int 14:19
Gunaratne A, Thai BL, Guglielmo GM (2013) Atypical protein kinase C phosphorylates Par6 and facilitates transforming growth factor beta-induced epithelial-to-mesenchymal transition. Mol Cell Biol 33:874–886
Mu Y, Zang G, Engstrom U, Busch C, Landstrom M (2015) TGFbeta-induced phosphorylation of Par6 promotes migration and invasion in prostate cancer cells. Brit J Cancer 112:1223–1231
Zhang K, Zhao H, Ji Z, Zhang C, Zhou P, Wang L et al (2016) Shp2 promotes metastasis of prostate cancer by attenuating the PAR3/PAR6/aPKC polarity protein complex and enhancing epithelial-to-mesenchymal transition. Oncogene 35:1271–1282
Barash U, Spyrou A, Liu P, Vlodavsky E, Zhu C, Luo J et al (2019) Heparanase promotes glioma progression via enhancing CD24 expression. Int J Cancer 145:1596–1608
Li Y, Wang H, Sun T, Chen J, Guo L, Shen H et al (2015) Biological characteristics of a new human glioma cell line transformed into A2B5+ stem cells. Mol Cancer 14:75
Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E et al (2012) IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483:479–483
Luo H, Chen Z, Wang S, Zhang R, Qiu W, Zhao L et al (2015) c-Myc-miR-29c-REV3L signalling pathway drives the acquisition of temozolomide resistance in glioblastoma. Brain 138:3654–3672
Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R et al (2009) Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 4:568–580
Duan JJ, Wang D, Cai J, Chen JJ, Zheng XX, Chen TQ et al (2022) An aldehyde dehydrogenase 1A3 inhibitor attenuates the metastasis of human colorectal cancer. Cancer Lett 536:215662
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Duan ZH, Wang HC, Zhao DM, Ji XX, Song M, Yang XJ et al (2015) Cooperatively transcriptional and epigenetic regulation of sonic hedgehog overexpression drives malignant potential of breast cancer. Cancer Sci 106:1084–1091
Liu P, Zhu C, Luo J, Lan S, Su D, Wang Q et al (2020) Par6 regulates cell cycle progression through enhancement of Akt/PI3K/GSK-3beta signaling pathway activation in glioma. FASEB J 34:1481–1496
Baghel KS, Tewari BN, Shrivastava R, Malik SA, Lone MU, Jain NK et al (2016) Macrophages promote matrix protrusive and invasive function of breast cancer cells via MIP-1beta dependent upregulation of MYO3A gene in breast cancer cells. Oncoimmunology 5:e1196299
Fan CC, Cheng WC, Huang YC, Sher YP, Liou NJ, Chien YC et al (2017) EFHD2 promotes epithelial-to-mesenchymal transition and correlates with postsurgical recurrence of stage I lung adenocarcinoma. Sci Rep 7:14617
Li Y, Zhang H, Gong H, Yuan Y, Li Y, Wang C et al (2018) miR-182 suppresses invadopodia formation and metastasis in non-small cell lung cancer by targeting cortactin gene. J Exp Clin Cancer Res 37:141
Jones ML, Siddiqui J, Pienta KJ, Getzenberg RH (2013) Circulating fibroblast-like cells in men with metastatic prostate cancer. Prostate 73:176–181
Xu X, Bao Z, Liu Y, Jiang K, Zhi T, Wang D et al (2018) PBX3/MEK/ERK1/2/LIN28/let-7b positive feedback loop enhances mesenchymal phenotype to promote glioblastoma migration and invasion. J Exp Clin Cancer Res 37:158
Balzeau J, Menezes MR, Cao S, Hagan JP (2017) The LIN28/let-7 pathway in cancer. Front Genet 8:31
Gunaratne A, Guglielmo GM (2013) Par6 is phosphorylated by aPKC to facilitate EMT. Cell Adhes Migr 7:357–361
Viloria-Petit AM, Wrana JL (2010) The TGFbeta-Par6 polarity pathway: linking the Par complex to EMT and breast cancer progression. Cell Cycle 9:623–624
Zagzag D, Salnikow K, Chiriboga L, Yee H, Lan L, Ali MA et al (2005) Downregulation of major histocompatibility complex antigens in invading glioma cells: stealth invasion of the brain. Lab Investig 85:328–341
Lefranc F, Brotchi J, Kiss R (2005) Possible future issues in the treatment of glioblastomas: special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J Clin Oncol 23:2411–2422
Annabi B, Lachambre MP, Plouffe K, Sartelet H, Beliveau R (2009) Modulation of invasive properties of CD133+ glioblastoma stem cells: a role for MT1-MMP in bioactive lysophospholipid signaling. Mol Carcinog 48:910–919
Merzak A, Koocheckpour S, Pilkington GJ (1994) CD44 mediates human glioma cell adhesion and invasion in vitro. Cancer Res 54:3988–3992
Okada H, Yoshida J, Sokabe M, Wakabayashi T, Hagiwara M (1996) Suppression of CD44 expression decreases migration and invasion of human glioma cells. Int J Cancer 66:255–260
Shi Q, Song X, Wang J, Gu J, Zhang W, Hu J et al (2015) FRK inhibits migration and invasion of human glioma cells by promoting N-cadherin/beta-catenin complex formation. J Mol Neurosci 55:32–41
Velpula KK, Rehman AA, Chelluboina B, Dasari VR, Gondi CS, Rao JS et al (2012) Glioma stem cell invasion through regulation of the interconnected ERK, integrin alpha6 and N-cadherin signaling pathway. Cell Signal 24:2076–2084
Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP et al (2006) Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol 8:1235–1245
Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T (2000) A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol 2:540–547
Butler AM, Scotti Buzhardt ML, Erdogan E, Li S, Inman KS, Fields AP et al (2015) A small molecule inhibitor of atypical protein kinase C signaling inhibits pancreatic cancer cell transformed growth and invasion. Oncotarget 6:15297–15310
Fang XY, Zhang H, Zhao L, Tan S, Ren QC, Wang L et al (2018) A new xanthatin analogue 1beta-hydroxyl-5alpha-chloro-8-epi-xanthatin induces apoptosis through ROS-mediated ERK/p38 MAPK activation and JAK2/STAT3 inhibition in human hepatocellular carcinoma. Biochimie 152:43–52
Fang Z, Tang Y, Fang J, Zhou Z, Xing Z, Guo Z et al (2013) Simvastatin inhibits renal cancer cell growth and metastasis via AKT/mTOR, ERK and JAK2/STAT3 pathway. PLoS One 8:e62823
Kim SJ, Pham TH, Bak Y, Ryu HW, Oh SR, Yoon DY (2018) Orientin inhibits invasion by suppressing MMP-9 and IL-8 expression via the PKCalpha/ ERK/AP-1/STAT3-mediated signaling pathways in TPA-treated MCF-7 breast cancer cells. Phytomedicine 50:35–42
Shi L, Wang S, Zangari M, Xu H, Cao TM, Xu C et al (2010) Over-expression of CKS1B activates both MEK/ERK and JAK/STAT3 signaling pathways and promotes myeloma cell drug-resistance. Oncotarget 1:22–33
Zhang D, Liu H, Yang B, Hu J, Cheng Y (2019) L-securinine inhibits cell growth and metastasis of human androgen-independent prostate cancer DU145 cells via regulating mitochondrial and AGTR1/MEK/ERK/STAT3/PAX2 apoptotic pathways. Biosci Rep 39:BSR20190469
Qin JJ, Yan L, Zhang J, Zhang WD (2019) STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review. J Exp Clin Cancer Res 38:195
Kim S, Kil WH, Lee J, Oh SJ, Han J, Jeon M et al (2014) Zerumbone suppresses EGF-induced CD44 expression through the inhibition of STAT3 in breast cancer cells. Oncol Rep 32:2666–2672
Wei B, Sun X, Geng Z, Shi M, Chen Z, Chen L et al (2016) Isoproterenol regulates CD44 expression in gastric cancer cells through STAT3/MicroRNA373 cascade. Biomaterials 105:89–101
Quintanal-Villalonga A, Ojeda-Marquez L, Marrugal A, Yague P, Ponce-Aix S, Salinas A et al (2018) The FGFR4-388arg variant promotes lung cancer progression by N-cadherin induction. Sci Rep 8:2394
Pei G, Lan Y, Chen D, Ji L, Hua ZC (2017) FAK regulates E-cadherin expression via p-SrcY416/p-ERK1/2/p-Stat3Y705 and PPARgamma/miR-125b/Stat3 signaling pathway in B16F10 melanoma cells. Oncotarget 8:13898–13908
Mu P, Liu K, Lin Q, Yang W, Liu D, Lin Z et al (2019) Sirtuin 7 promotes glioma proliferation and invasion through activation of the ERK/STAT3 signaling pathway. Oncol Lett 17:1445–1452
Horvitz HR, Sulston JE (1980) Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96:435–454
Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD et al (2017) H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis 8:e2569
Shyh-Chang N, Daley GQ (2013) Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 12:395–406
Wang T, Wang G, Hao D, Liu X, Wang D, Ning N et al (2015) Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer. Mol Cancer 14:125
Newman MA, Thomson JM, Hammond SM (2008) Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14:1539–1549
Dong Q, Meng P, Wang T, Qin W, Qin W, Wang F et al (2010) MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS One 5:e10147
Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P et al (2007) MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 67:9762–9770
Wang XR, Luo H, Li HL, Cao L, Wang XF, Yan W et al (2013) Overexpressed let-7a inhibits glioma cell malignancy by directly targeting K-ras, independently of PTEN. Neuro-Oncology 15:1491–1501
Han L, Wang Y, Wang L, Guo B, Pei S, Jia Y (2018) MicroRNA let-7f-5p regulates neuronal differentiation of rat bone marrow mesenchymal stem cells by targeting Par6alpha. Biochem Biophys Res Commun 495:1476–1481
Guo Z, Li G, Bian E, Ma CC, Wan J, Zhao B (2017) TGF-beta-mediated repression of MST1 by DNMT1 promotes glioma malignancy. Biomed Pharmacother 94:774–780
Singh SK, Fiorelli R, Kupp R, Rajan S, Szeto E, Lo Cascio C et al (2016) Post-translational modifications of OLIG2 regulate glioma invasion through the TGF-beta pathway. Cell Rep 16:950–966
Bayin NS, Ma L, Thomas C, Baitalmal R, Sure A, Fansiwala K et al (2016) Patient-specific screening using high-grade glioma explants to determine potential radiosensitization by a TGF-beta small molecule inhibitor. Neoplasia 18:795–805
Joseph JV, Conroy S, Tomar T, Eggens-Meijer E, Bhat K, Copray S et al (2014) TGF-beta is an inducer of ZEB1-dependent mesenchymal transdifferentiation in glioblastoma that is associated with tumor invasion. Cell Death Dis 5:e1443
Mahabir R, Tanino M, Elmansuri A, Wang L, Kimura T, Itoh T et al (2014) Sustained elevation of Snail promotes glial-mesenchymal transition after irradiation in malignant glioma. Neuro-Oncology 16:671–685
Zheng Y, Miu Y, Yang X, Yang X, Zhu M (2017) CCR7 mediates TGF-beta1-induced human malignant glioma invasion, migration, and epithelial-mesenchymal transition by activating MMP2/9 through the nuclear factor KappaB signaling pathway. DNA Cell Biol 36:853–861
Acknowledgements
Not applicable.
Funding
This study was supported by National Natural Science Foundation of China (81872070 and 81673652), Science and Technique Foundation of Guangdong Province (210728156901639), and Natural Science Foundation of Guangdong Province (2022A1515012424).
Author information
Authors and Affiliations
Contributions
YH, PL, JL, CZ, CL, and NZ performed experiments and collected data. WZ and WC reviewed and helped the manuscript writing. WC and XY supervised the project, and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
All animal experiments in this study were approved by the Shantou University Medical College Animal Committee. Ethical approvals were from the Medical Ethics Committee of Shantou University Medical College (no. SUMC2019-002 and SUMC2020-73).
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information

Fig. S1
The inhibition of MEK/ERK signaling pathways significantly inhibits Par6-mediated glioma invasion. (A,B) The determination of wound healing (A) and transwell assay (B) in Par6-OE with or without U0126 treatment (n = 3 for each group). Scale bars, 200 μm. (PNG 2345 kb)

Fig. S2
TGF-β treatment induces glioma invasion through upregulating Par6 expression. (A) Par6 expression in TGF-β stimulation in different time points (0, 1, 2, 4, and 8 h) in U87MG cells. (B) TGF-β stimulation induces the upregulation of Par6 in time-dependent manner from 12 to 48 h in U87MG and U251 cells. (C) The expression of Par6, CD44, and N-cadherin in TGF-β-treated U251 and U87MG cells. (D) The expression of MEK, pMEK, ERK1/2, and pERK1/2 in U87MG and U251 cells with or without TGF-β treatment. (E) The expression of STAT3, pSTAT3 in U251 and U87MG cells with or without TGF-β treatment. (F) The representative images and quantitative analyses of glioma invasion in U251 and U87MG cells with or without TGF-β treatment, respectively (n = 3 for each group). Scale bar, 200 μm. (G) AT-1 treatment inhibit TGF-β-stimulated glioma invasion in U87MG cells. Scale bar, 200 μm. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, no significance. (PNG 802 kb)

Fig. S3
The Par6 expression is positively correlative with the glioma invasion and migration in primary glioma cells. (A) The expression levels of CD44, N-cadherin, MMP2, and MMP9 in Par6-OE, Par6-KD, and control groups of the cell lines from two primary glioma specimens (GBM1 and GBM2). (B) Cell invasion assays in Par6-OE, Par6-KD and control groups in two primary glioma cell lines (n = 3 random fields of view each group). Representative images of invading cells visualized by crystal violet staining, and quantification of cell invading capacity at 24 h. Scale bar, 200 μm. (C) Wound healing for cell migration in Par6-OE, Par6-KD and control groups in primary glioma cells. Representative images and quantification of cell migration into the wounded area at 0, 12, and 24 h (n = 3 for each group). Scale bar, 200 μm. *P < 0.05, **P < 0.01, ***P < 0.001. (PNG 1264 kb)

Fig. S4
The validation of downstream LIN28/let-7d axis in Par6-mediated tumor invasion in primary glioma cells. (A) Western blot determination and quantification of LIN28 expression in different groups in primary glioma cells. (B,C) Wound healing and transwell assays for migration (B) and invasion (C) in Par6-OE groups in primary glioma cells with or without the inhibition of LIN28 expression (n = 3 for each group). (D) The downregulation of CD44 and N-cadherin expression in Par6-OE cells after the inhibition of LIN28 expression. (E) qPCR assays were performed to determine the levels of let-7d in Par6-OE and control cells. (F) U0126 treatment can induce the upregulation of let-7d in Par6-OE groups in primary glioma cells. (G,H) Wound healing and transwell assays for migration (G) and invasion (H) in Par6-KD groups in primary glioma cells with or without the treatment of let-7d inhibitor (n = 3 for each group). (I,J) Western blot determination and quantification of CD44, N-cadherin (I), STAT3, and pSTAT3 (J) in Par6-KD groups in primary glioma cells with or without the treatment of let-7d inhibitor. *P < 0.05, **P < 0.01, ***P < 0.001. (PNG 1179 kb)
Supplementary file 1
(DOCX 25 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, Y., Liu, P., Luo, J. et al. Par6 Enhances Glioma Invasion by Activating MEK/ERK Pathway Through a LIN28/let-7d Positive Feedback Loop. Mol Neurobiol 60, 1626–1644 (2023). https://doi.org/10.1007/s12035-022-03171-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-03171-0