Abstract
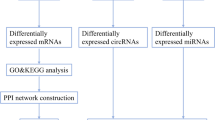
Emerging evidence suggested that long non-coding RNAs (lncRNAs) were involved in Parkinson’s disease (PD) pathogenesis. Herein, we used gene expression profiles from GEO database to construct a PD-specific ceRNA network. Functional enrichment analysis suggested that ceRNA network might participate in the development of PD. PPI networks were constructed, and the ceRNA subnetwork based on five hub genes was set up. In a cohort of 32 PD patients and 31 healthy controls, the expression of 10 DElncRNAs (TTC3-AS1, LINC01259, ZMYND10-AS1, CHRM3-AS1, MYO16-AS1, AGBL5-IT1, HOTAIRM1, RABGAP1L-IT1, HLCS-IT1, and LINC00393) were further verified. Consistent with the microarray data, LINC01259 expression was significantly lower in PD patients compared with controls (P = 0.008). Intriguingly, such a difference was only observed among male patients and male controls when dividing study participants based on their gender (P = 0.016). However, the expression of other lncRNAs did not differ significantly between the two groups. Receiver operating characteristic (ROC) curve analysis revealed that the diagnostic power of LINC01259 was 0.694 for PD and 0.677 for early-stage PD. GSEA enrichment analysis revealed that LINC01259 was mainly enriched in biological processes associated with immune function and inflammatory response. Moreover, LINC01259 expression was not correlated with age of patients, disease duration, disease stage, MDS-UPDRS score, MDS-UPDRS III score, MMSE score, and MOCA score. The current study provides further evidence for the dysregulation of lncRNAs in circulating leukocytes of PD patients, revealing that LINC01259 has clinical potential as a novel immune and inflammatory biomarker for PD and early-stage PD diagnosis.







Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, Schrag AE, Lang AE (2017) Parkinson disease Nat Rev Dis Primers 3:17013. https://doi.org/10.1038/nrdp.2017.13
Fayyad M, Salim S, Majbour N, Erskine D, Stoops E, Mollenhauer B, El-Agnaf OMA (2019) Parkinson’s disease biomarkers based on α-synuclein. J Neurochem 150:626–636. https://doi.org/10.1111/jnc.14809
Bloem BR, Okun MS, Klein C (2021) Parkinson’s disease. Lancet. https://doi.org/10.1016/s0140-6736(21)00218-x
Jankovic J (2008) Parkinson’s disease: clinical features and diagnosis. J Neurol Neurosurg Psychiatry 79:368–376. https://doi.org/10.1136/jnnp.2007.131045
Barone P, Erro R, Picillo M (2017) Quality of life and nonmotor symptoms in Parkinson’s disease. Int Rev Neurobiol 133:499–516. https://doi.org/10.1016/bs.irn.2017.05.023
Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12:861–874. https://doi.org/10.1038/nrg3074
Statello L, Guo CJ, Chen LL, Huarte M (2021) Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22:96–118. https://doi.org/10.1038/s41580-020-00315-9
Schmitz SU, Grote P, Herrmann BG (2016) Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci 73:2491–2509. https://doi.org/10.1007/s00018-016-2174-5
Ng SY, Lin L, Soh BS, Stanton LW (2013) Long noncoding RNAs in development and disease of the central nervous system. Trends Genet 29:461–468. https://doi.org/10.1016/j.tig.2013.03.002
Khorkova O, Hsiao J, Wahlestedt C (2015) Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev 87:15–24. https://doi.org/10.1016/j.addr.2015.05.012
Liu Y, Lu Z (2018) Long non-coding RNA NEAT1 mediates the toxic of Parkinson’s disease induced by MPTP/MPP+ via regulation of gene expression. Clin Exp Pharmacol Physiol 45:841–848. https://doi.org/10.1111/1440-1681.12932
Lin Q, Hou S, Dai Y, Jiang N, Lin Y (2019) LncRNA HOTAIR targets miR-126-5p to promote the progression of Parkinson’s disease through RAB3IP. Biol Chem 400:1217–1228. https://doi.org/10.1515/hsz-2018-0431
Chen Y, Lian YJ, Ma YQ, Wu CJ, Zheng YK, Xie NC (2018) LncRNA SNHG1 promotes α-synuclein aggregation and toxicity by targeting miR-15b-5p to activate SIAH1 in human neuroblastoma SH-SY5Y cells. Neurotoxicology 68:212–221. https://doi.org/10.1016/j.neuro.2017.12.001
Xu X, Zhuang C, Wu Z, Qiu H, Feng H, Wu J (2018) LincRNA-p21 inhibits cell viability and promotes cell apoptosis in Parkinson’s disease through activating α-synuclein expression. Biomed Res Int 2018:8181374. https://doi.org/10.1155/2018/8181374
Song Z, Xie B (2021) LncRNA OIP5-AS1 reduces α-synuclein aggregation and toxicity by targeting miR-126 to activate PLK2 in human neuroblastoma SH-SY5Y cells. Neurosci Lett 740:135482. https://doi.org/10.1016/j.neulet.2020.135482
Calabrese V, Santoro A, Monti D, Crupi R, Di Paola R, Latteri S, Cuzzocrea S, Zappia M, et al. (2018) Aging and Parkinson’s disease: inflammaging, neuroinflammation and biological remodeling as key factors in pathogenesis. Free Radic Biol Med 115:80–91. https://doi.org/10.1016/j.freeradbiomed.2017.10.379
Tansey MG, Wallings RL, Houser MC, Herrick MK, Keating CE, Joers V (2022) Inflammation and immune dysfunction in Parkinson disease. Nat Rev Immunol 1-17. https://doi.org/10.1038/s41577-022-00684-6
Ferrari CC, Tarelli R (2011) Parkinson’s disease and systemic inflammation. Parkinsons Dis 2011:436813. https://doi.org/10.4061/2011/436813
Zhang W, Wang T, Pei Z, Miller DS, Wu X, Block ML, Wilson B, Zhang W, et al. (2005) Aggregated alpha-synuclein activates microglia: a process leading to disease progression in Parkinson’s disease. Faseb j 19:533–542. https://doi.org/10.1096/fj.04-2751com
Tecchio C, Micheletti A, Cassatella MA (2014) Neutrophil-derived cytokines: facts beyond expression. Front Immunol 5:508. https://doi.org/10.3389/fimmu.2014.00508
Liu X, Quan N (2018) Microglia and CNS interleukin-1: beyond immunological concepts. Front Neurol 9:8. https://doi.org/10.3389/fneur.2018.00008
Jensen MP, Jacobs BM, Dobson R, Bandres-Ciga S, Blauwendraat C, Schrag A, Noyce AJ (2021) Lower lymphocyte count is associated with increased risk of Parkinson’s disease. Ann Neurol 89:803–812. https://doi.org/10.1002/ana.26034
Akıl E, Bulut A, Kaplan İ, Özdemir HH, Arslan D, Aluçlu MU (2015) The increase of carcinoembryonic antigen (CEA), high-sensitivity C-reactive protein, and neutrophil/lymphocyte ratio in Parkinson’s disease. Neurol Sci 36:423–428. https://doi.org/10.1007/s10072-014-1976-1
Muñoz-Delgado L, Macías-García D, Jesús S, Martín-Rodríguez JF, Labrador-Espinosa M, Jiménez-Jaraba MV, Adarmes-Gómez A, Carrillo F, Mir P (2021) Peripheral immune profile and neutrophil-to-lymphocyte ratio in Parkinson’s disease. Mov Disord 36:2426–2430. https://doi.org/10.1002/mds.28685
Shi Y, Wei B, Li L, Wang B, Sun M (2022) Th17 cells and inflammation in neurological disorders: possible mechanisms of action. Front Immunol 13:932152. https://doi.org/10.3389/fimmu.2022.932152
Contaldi E, Magistrelli L, Comi C (2022) T lymphocytes in Parkinson’s disease. J Parkinsons Dis. https://doi.org/10.3233/jpd-223152
Runtsch MC, Ferrara G, Angiari S (2021) Metabolic determinants of leukocyte pathogenicity in neurological diseases. J Neurochem 158:36–58. https://doi.org/10.1111/jnc.15169
Heward JA, Lindsay MA (2014) Long non-coding RNAs in the regulation of the immune response. Trends Immunol 35:408–419. https://doi.org/10.1016/j.it.2014.07.005
Mowel WK, Kotzin JJ, McCright SJ, Neal VD, Henao-Mejia J (2018) Control of Immune cell homeostasis and function by lncRNAs. Trends Immunol 39:55–69. https://doi.org/10.1016/j.it.2017.08.009
Li G, Ma X, Zhao H, Fan J, Liu T, Luo Y, Guo Y (2022) Long non-coding RNA H19 promotes leukocyte inflammation in ischemic stroke by targeting the miR-29b/C1QTNF6 axis. CNS Neurosci Ther 28:953–963. https://doi.org/10.1111/cns.13829
Soreq L, Guffanti A, Salomonis N, Simchovitz A, Israel Z, Bergman H, Soreq H (2014) Long non-coding RNA and alternative splicing modulations in Parkinson’s leukocytes identified by RNA sequencing. PLoS Comput Biol 10:e1003517. https://doi.org/10.1371/journal.pcbi.1003517
Fan Y, Li J, Yang Q, Gong C, Gao H, Mao Z, Yuan X, Zhu S, et al. (2019) Dysregulated long non-coding RNAs in Parkinson’s disease contribute to the apoptosis of human neuroblastoma cells. Front Neurosci 13:1320. https://doi.org/10.3389/fnins.2019.01320
Yang P, Lin G, Wang M, Chen X, Huang J (2022) Long non-coding RNA ANRIL interacts with microRNA-34a and microRNA-125a, and they all correlate with disease risk and severity of Parkinson’s disease. J Clin Lab Anal 36:e24037. https://doi.org/10.1002/jcla.24037
Yang X, Zhang Y, Chen Y, He X, Qian Y, Xu S, Gao C, Mo C, et al. (2021) LncRNA HOXA-AS2 regulates microglial polarization via recruitment of PRC2 and epigenetic modification of PGC-1α expression. J Neuroinflammation 18:197. https://doi.org/10.1186/s12974-021-02267-z
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, et al. (2013) NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 41:D991–D995. https://doi.org/10.1093/nar/gks1193
Davis S, Meltzer PS (2007) GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23:1846–1847. https://doi.org/10.1093/bioinformatics/btm254
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. https://doi.org/10.1093/nar/gkv007
Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH (2011) starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res 39:D202–D209. https://doi.org/10.1093/nar/gkq1056
Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4. https://doi.org/10.7554/eLife.05005
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, et al. (2011) miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res 39:D163–D169. https://doi.org/10.1093/nar/gkq1107
Chen Y, Wang X (2020) miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 48:D127-d131. https://doi.org/10.1093/nar/gkz757
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. https://doi.org/10.1089/omi.2011.0118
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, et al. (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607-d613. https://doi.org/10.1093/nar/gky1131
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8(Suppl 4):S11. https://doi.org/10.1186/1752-0509-8-s4-s11
Postuma RB, Berg D, Stern M, Poewe W, Olanow CW, Oertel W, Obeso J, Marek K, et al. (2015) MDS clinical diagnostic criteria for Parkinson’s disease. Mov Disord 30:1591–1601. https://doi.org/10.1002/mds.26424
Goetz CG, Tilley BC, Shaftman SR, Stebbins GT, Fahn S, Martinez-Martin P, Poewe W, Sampaio C, et al. (2008) Movement Disorder Society-sponsored revision of the Unified Parkinson’s Disease Rating Scale (MDS-UPDRS): scale presentation and clinimetric testing results. Mov Disord 23:2129–2170. https://doi.org/10.1002/mds.22340
P. W (2012) Global scales to stage disability in PD: the Hoehn and Yahr scale. Rating Scales Parkinsons Dis 115–22. https://doi.org/10.1093/med/9780199783106.003.0258
Arevalo-Rodriguez I, Smailagic N, Roqué IFM, Ciapponi A, Sanchez-Perez E, Giannakou A, Pedraza OL, Bonfill Cosp X et al. (2015) Mini-Mental State Examination (MMSE) for the detection of Alzheimer’s disease and other dementias in people with mild cognitive impairment (MCI). Cochrane Database Syst Rev 2015:Cd010783. https://doi.org/10.1002/14651858.CD010783.pub2
Nasreddine ZS, Phillips NA, Bédirian V, Charbonneau S, Whitehead V, Collin I, Cummings JL, Chertkow H (2005) The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc 53:695–699. https://doi.org/10.1111/j.1532-5415.2005.53221.x
Kraus TFJ, Haider M, Spanner J, Steinmaurer M, Dietinger V, Kretzschmar HA (2017) Altered long noncoding RNA expression precedes the course of Parkinson’s disease-a preliminary report. Mol Neurobiol 54:2869–2877. https://doi.org/10.1007/s12035-016-9854-x
Coupland KG, Kim WS, Halliday GM, Hallupp M, Dobson-Stone C, Kwok JB (2016) Role of the Long Non-Coding RNA MAPT-AS1 in Regulation of Microtubule Associated Protein Tau (MAPT) Expression in Parkinson’s Disease. PLoS ONE 11:e0157924. https://doi.org/10.1371/journal.pone.0157924
Moreno-García L, López-Royo Y, Calvo AC, Toivonen JM, de la Torre M, Moreno-Martínez L, Molina N, Aparicio P, Zaragoza P, et al. (2020) Competing endogenous RNA networks as biomarkers in neurodegenerative diseases. Int J Mol Sci 21. https://doi.org/10.3390/ijms21249582
Zhang X, Feng S, Fan Y, Luo Y, Jin L, Li S (2020) Identifying a comprehensive ceRNA network to reveal novel targets for the pathogenesis of Parkinson’s disease. Front Neurol 11:810. https://doi.org/10.3389/fneur.2020.00810
Zhao J, Geng L, Chen Y, Wu C (2020) SNHG1 promotes MPP(+)-induced cytotoxicity by regulating PTEN/AKT/mTOR signaling pathway in SH-SY5Y cells via sponging miR-153-3p. Biol Res 53:1. https://doi.org/10.1186/s40659-019-0267-y
Chen Q, Huang X, Li R (2018) lncRNA MALAT1/miR-205-5p axis regulates MPP(+)-induced cell apoptosis in MN9D cells by directly targeting LRRK2. Am J Transl Res 10:563–572
Cao B, Wang T, Qu Q, Kang T, Yang Q (2018) Long noncoding RNA SNHG1 promotes neuroinflammation in Parkinson’s disease via regulating miR-7/NLRP3 pathway. Neuroscience 388:118–127. https://doi.org/10.1016/j.neuroscience.2018.07.019
Zhu Z, Huang P, Sun R, Li X, Li W, Gong W (2022) A novel long-noncoding RNA LncZFAS1 prevents MPP(+)-induced neuroinflammation through MIB1 activation. Mol Neurobiol 59:778–799. https://doi.org/10.1007/s12035-021-02619-z
Kim EK, Choi EJ (2015) Compromised MAPK signaling in human diseases: an update. Arch Toxicol 89:867–882. https://doi.org/10.1007/s00204-015-1472-2
Cerri S, Mus L, Blandini F (2019) Parkinson’s Disease in women and men: what’s the difference? J Parkinsons Dis 9:501–515. https://doi.org/10.3233/jpd-191683
Acknowledgements
We would like to thank Dr. Jiahang Song and Dr. Yifang Hu for the experimental guidance and useful discussion of the work.
Funding
This work was supported by the Natural Science Foundation of Jiangsu Province (BK20201117) and the National Science and Technology Innovation 2030: Major program of “Brain Science and Brain-Inspired Intelligence Research” (2021ZD0201807).
Author information
Authors and Affiliations
Contributions
Study design and manuscript draft: YDZ, YYT, and TH; experiment implementation: TH, JYZ, RRP, and TJ; data collection: XXF, QH, XXW, and PYG. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
All the participants gave written informed consent. The research on human subjects was performed in accordance with the ethical standards of the Declaration of Helsinki and approved by the ethics committee of the Nanjing First Hospital (ethical permits KY20220124-06).
Consent for Publication
All the data is suitable for publication.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, T., Zhao, JY., Pan, RR. et al. Dysregulation of Circulatory Levels of lncRNAs in Parkinson’s Disease. Mol Neurobiol 60, 317–328 (2023). https://doi.org/10.1007/s12035-022-03086-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-03086-w