Abstract
The pathological hallmark of the majority of amyotrophic lateral sclerosis (ALS) cases is the mislocalization and aggregation of TAR DNA-binding protein 43 (TDP-43), an RNA-binding protein. Several studies have attributed disease processes of ALS to abnormal RNA metabolism. However, dysregulated biogenesis of RNA, especially non-coding RNA (ncRNA), is poorly understood. To resolve it, RNA-Seq, biochemical, and immunohistochemical analyses were performed on the pyramidal tract of the medulla oblongata of sporadic ALS (sALS) and control postmortem brain samples. Here, we report perturbation of ncRNA biogenesis in PIWI-interacting RNA (piRNA) in several sALS brain samples associated with TDP-43 pathology. In addition, we confirmed the dysregulation of two PIWI homologs, PIWI-like-mediated gene silencing 1 (PIWIL1) and PIWIL4, which bind to piRNAs to regulate their expression. PIWIL1 was mislocalized and co-localized with TDP-43 in motor neurons of sporadic ALS lumbar cords. Our results imply that dysregulation of piRNA, PIWIL1, and PIWIL4 is linked to pathogenesis of ALS. Based on these results, piRNAs and PIWI proteins are potential diagnostic biomarkers and therapeutic targets of ALS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During the last decade, significant progress has been made in identifying genes (~ 50) responsible for familial amyotrophic lateral sclerosis (fALS) [1]. However, fALS constitutes only 10% of all ALS cases. The remaining 90% are termed “sporadic” ALS, and most of these cases have no known genetically inherited component. The lack of an identifiable cause of these sporadic cases makes it difficult to diagnose ALS early enough to distinguish it from other diseases causing similar symptoms.
TAR DNA-binding protein 43 (TDP-43) is a highly conserved and ubiquitously expressed RNA-binding protein, which belongs to the heterogeneous nuclear ribonucleoprotein (hnRNP) family [2]. One of the hallmarks of ALS in the vast majority of both familial and sporadic ALS cases is mislocalization, aggregation, and inclusion formation of TDP-43 protein in brain tissues. The function of TDP-43 strongly suggests its role in RNA metabolism of coding and non-coding RNAs (ncRNAs), including small ncRNAs. Apart from fALS, the only possible way to study the pathogenesis of sporadic ALS (sALS) is to compare patient samples to control samples in order to discover potential gene regulatory targets, which should be beneficial for establishing biomarkers to diagnose the disease and to develop treatment strategies to block their aberrant expression.
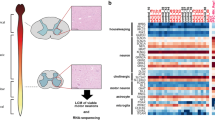
Based on these findings, we conducted a pilot RNA-seq study using postmortem brain samples of sALS patients and controls with no known neurological disease (Fig. 1). Differential expression analysis revealed dysregulation in small ncRNAs, particularly in micro RNAs (miRNAs) and PIWI-interacting RNA (piRNA). Owing to recent reports of potential involvement of piRNAs in the etiology of different neurological diseases, such as Rett syndrome, Alzheimer’s disease (AD), Parkinson’s disease (PD) [3], and brain aging [4], we focused on piRNA dysregulation in sALS samples to identify targets of those piRNAs and to determine whether PIWI protein/piRNAs might contribute to ALS pathology.
Schematic representation of the experimental design. Postmortem human tissue samples used in this study were collected from ALS cases (n = 7) and age- and sex-matched control cases (n = 7). RNA-Seq was performed on RNA extracted from the pyramidal tract of the medulla oblongata. Differential expression analysis of small RNAs was performed, followed by validation
Methods
Postmortem Brain Samples
Postmortem human tissue samples used in this study were collected from the Brain Bank for Aging Research (BBAR) at the Tokyo Metropolitan Geriatric Hospital and Institute of Gerontology (TMGHIG), and the National Center of Neurology and Psychiatry (NCNP) Brain Bank. sALS cases (n = 7) were diagnosed based on El Escorial and Airlie House revised criteria with no familial history of the disease and confirmed pathological inclusions of TDP-43 in brain lesions. The stage of TDP-43 pathology that determines the severity of ALS was defined according to the criteria by Brettschneider et al. [5]. Age- and sex-matched control cases (n = 7) had no neurological pathologies (Table 1). The pyramidal tract of the medulla oblongata was chosen for this study because this area is rich in axons of upper motor neurons, which are affected in ALS.
cDNA Preparation for Small RNA-Seq
Total RNA used for RNA-seq was extracted using Isogen-LS (Nippon Gene # 311–02,501). RNA concentration was measured with a Qubit RNA Assay Kit in a Qubit 2.0 Fluorometer (Life Technologies, CA, USA). RNA integrity was evaluated with an RNA Pico 6000 Assay Kit and a Bioanalyzer 2100 System (Agilent Technologies, CA, USA). The mean RIN for ALS samples was 7.9 (range 6.9–8.5). 50 ng of each RNA sample was used to construct a cDNA library for each sample using a NEXTFLEX Combo-Seq mRNA/miRNA kit (Bio-Scientific), according to the manufacturer’s protocol (Nova-5139–01). cDNAs were sequenced on an Illumina HiSeq 3000 with 74-bp single reads (10 M reads/sample).
RNA-Seq Bioinformatic Analysis
We excluded adapter sequences using the cutadapt tool V.1.9.2 [6]. Reads shorter than 15 bases were eliminated using trimmomatic version 0.36 [7]. Then, we conducted comprehensive small RNA-seq analysis using the exceRpt extra-cellular RNA processing toolkit, exceRpt version 4.6.3 (https://rkitchen.github.io/exceRpt/) [8].
First, exceRpt automatically excludes low-quality reads. Next, reads aligned to sequences in the Univec Vecscreen database and human ribosomal RNA sequences are removed to filter out contaminants. After filtering, high-quality reads are aligned to the host transcriptome and genome and transcriptome abundances are calculated. In this package, miRbase, circBase, GtRNAdb, piRNABank, and GENCODE are used to assign reads to miRNAs, circular RNAs, tRNAs, piRNAs, and longRNAs.
miRbase (https://mirbase.org/)
circBase (http://circbase.org/)
GtRNAdb (http://gtrnadb.ucsc.edu/)
piRNABank (http://pirnabank.ibab.ac.in/)
GENCODE (https://www.gencodegenes.org/)
Data Resources
Raw FASTQ files for the RNA-seq libraries have been submitted to the DNA Data Bank of Japan (DDBJ) with Temporary Submission ID: SSUB016807.
RNA Extraction for Reverse Transcription-Quantitative PCR (RT-qPCR) Validation
Total RNA was extracted from medulla oblongata samples using a mirVana kit (Ambion), according to manufacturer’s instructions. Small RNA (sRNA) was isolated from total RNA using the mirVana kit (Ambion), according to the manufacturer’s instructions for sRNA isolation.
RT-qPCR for piRNAs
In brief, 3′-ends of 60 ng of sRNA from each sample were ligated to 5′ pre-adenylated 3′-adaptors (5′- rApp/CTGTAGGCACCATCAAT/3ddC-3′) using truncated T4RNA ligase 2 enzyme (NEB). 3′- adaptor-ligated sRNAs were reverse transcribed with ReverTraAce, reverse transcription polymerase (Toyobo) using an oligonucleotide complementary to the 3′-adaptor (IDT) in a MiniAmp plus Thermal Cycler (Applied Biosystems). RT-qPCR was performed using forward primers for each target piRNA and the oligonucleotide complementary to the 3′-adaptor was used as a reverse primer (Table 5). This technique only allows the detection of mature piRNAs. Data were collected in triplicate for each sample on an ABI 7900 Prism qPCR machine and normalized using U6RNA as an internal control. Relative gene-expression levels were calculated using the fold-change method.
RT-qPCR for Coding Genes
RT-qPCR was performed using ReverTraAce (Toyobo) in a MiniAmp plus Thermal Cycler (Applied Biosystems). The glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) was used as an internal control to normalize coding genes. All quantitative PCR was performed using SYBRGreen qPCR Master Mix (Applied Biosystems) on an ABI 7900 Prism qPCR machine. Primer sets used for RT-qPCR are listed in Table 5.
Standard Curve Method for RT-qPCR
We used the standard curve method to calculate the average fold change of piRNA expression in ALS samples comparing to control samples.
cDNA generated by reverse transcription for seven controls and seven ALS samples were used for the fold-change calculation. Equal volumes of control cDNA were pooled as a control stock. The control stock was then diluted 10 times with nuclease-free water in a dilution series to produce a control standard curve. sALS cDNA samples were diluted 20 times and then plotted against the control standard curve for each gene in triplicate. GAPDH was used as an internal control for normalization.
△CT Method for RT-qPCR
We used ΔCT method to study piRNA expression of each sample (control and ALS) individually.
The △CT RT-qPCR method was used to calculate the difference in cycle numbers needed to amplify target genes after normalization using the internal control. Therefore, △CT denotes the CT value of a target gene subtracted by the CT value of the internal control. Internal controls for piRNA and coding genes were U6RNA and GAPDH, respectively.
Tissue Lysates and Protein Quantification
CellLytic MT Mammalian Tissue Lysis Reagent (C3228, Sigma-Aldrich), mixed with a protease inhibitor cocktail (1:100, EMD Millipore), was added to frozen tissue in 2-mL Lysing Matrix D tubes (P000912-LYSK0, Precellys) to be homogenized using a MINILYS personal homogenizer (Bretin Instruments). Homogenized tissue lysates were centrifuged and supernatants were used for protein concentration determinations with a Pierce TM BCA protein assay (23225, Thermo Fisher Scientific).
Immunoblotting for PIWI Proteins
Thirty micrograms of protein from lysates was separated on 5–20% SDS–PAGE gels (E-T520L, ATTO) and transferred to 0.2-μm PVDF membranes (1620177, Bio-Rad). Membranes were blocked for 1 h in EZ blocking solution (AE-1475, ATTO) and then incubated with primary antibodies diluted in Can Get Signal Solution 1 (TOYOBO) overnight at 4 °C. Primary antibodies used included PIWIL1 (1:1000; #701177, Novex), PIWIL2 (1:500; ab181340, Abcam), PIWIL3 (1:500; ab77088, Abcam), PIWIL4 (1:1000; ab111714, Abcam), PIWIL4 (1:1000; PA-49710, Thermo Fisher Scientific), and TXRND1 (1:1000; ab16840, Abcam).
After primary antibody incubation, membranes were washed with 0.1% T-TBS buffer three times (for 15, 10, and 5 min each) before incubation with secondary antibody. Secondary antibodies were HRP anti-mouse antibody (1:10000; NA931V, GE Healthcare) or anti-rabbit antibody (1:20000; NA934V, GE Healthcare) diluted in Can get Signal solution2 (TOYOBO) or EZ block solution and supplemented with STREP-TACTIN (#1616380, Bio-Rad) for marker band detection. After secondary antibody incubation, membranes were washed with 0.1% T-TBS buffer 3 × 10 min before visualization.
Data Analysis and Statistics (Immunoblotting and RT-qPCR)
Statistics were performed using GraphPad Prism, version9 software. Two-way ANOVA was performed with Bonferroni’s multiple comparison test to compare two or more independent groups. Pairwise comparisons were made using the Mann–Whitney test for △CT with unpaired and nonparametric settings. Pairwise comparisons for fold change using the standard curve method were made using one-sample t-tests (one-tailed Wilcoxon test) with paired and nonparametric settings.
For immunoblot statistical analysis, two-way ANOVA was performed using Bonferroni’s multiple comparison test to compare two or more proteins. Pairwise comparisons were made using the Mann–Whitney test for PIWI proteins with unpaired and nonparametric, two-tailed p-value settings. p-values < 0.05 were considered significant (* indicates p < 0.05 and ** p < 0.01). All numbers in plots represent means ± SEM.
Neuropathological Examination
Clinical profiles of patients examined in the present study are shown in Supplementary Table 1. Samples from the lumbar cord (L5) were fixed in 10% buffered formalin. For immunohistochemistry (IHC), 6-µm sections were prepared. Deparaffinized sections were incubated 30 min with 0.3% hydrogen peroxide to quench endogenous peroxidase activity and then washed with PBS. The primary antibody was a mouse monoclonal antibody against PIWIL1 (1:500). Samples were autoclaved 15 min before incubation with antibody. Secondary antibody was goat anti-mouse immunoglobulin conjugated to peroxidase-labeled dextran polymer (Dako Envision + , Dako). Reaction products were visualized with 3,3'-diaminobenzidine tetrahydrochloride (ImmPACT DAB, Vector Laboratories), and hematoxylin was used to counterstain cell nuclei. For double IHC, two primary antibodies were combined, including antibodies against TDP-43 (1:1000, rabbit polyclonal, Proteintech) and PIWIL1 (1:500). Alexa Fluor® 488 goat anti-mouse IgG (H + L) antibody (A-11008, Thermo Fisher Scientific) and Alexa Fluor® 568 goat anti-rabbit IgG (H + L) antibody (A-11004, Thermo Fisher Scientific) were used as secondary antibodies. Sudan Black B treatment was performed to reduce autofluorescence from lipofuscin. Images were obtained using an all-in-one fluorescence microscope (BZ-X710, Keyence).
Results
RNA-Seq Analysis for sALS Postmortem Samples
cDNA libraries were prepared using a Combo-Seq kit from seven control and seven sALS postmortem samples (Table 1). This kit was used to analyze poly A-tailed RNA and small ncRNA at the same time. However, it was difficult to get reliable annotation for poly A-tailed RNA during the bioinformatic analysis due to short reads, so we focused on analyzing only reads for small RNA-seq output. Data from two controls (CTR-1 and CTR-7) and sALS (ALS-1 and ALS-3) were excluded from the analysis due to low read numbers obtained from those libraries. Therefore, differential expression analysis was done using five controls and five sALS samples.
The analysis revealed significant dysregulation (p < 0.05) of nine miRNAs in ALS samples compared to controls (Fig. 2A; Table 2). Among dysregulated miRNAs, hsa-miR-143-5p and hsa-miR-143-3p [9,10,11] were previously reported as dysregulated in cerebrospinal fluid (CSF), serum, and immortalized lymphoblast cell lines (LCLs) derived from ALS patient samples, validating parameters used for data analysis in this study.
miRNAs and piRNAs are dysregulated in sALS. A miRNAs are dysregulated in sALS. Differential expression analysis revealed that nine miRNAs were significantly dysregulated in ALS samples (p < 0.05). Data were normalized as the log 10 read count per million (RPM). B piRNAs are significantly dysregulated in sALS. Three piRNAs were up-regulated in ALS samples (red) and two were down-regulated (blue) (p < 0.05). Data were normalized as the log 10 read count per million (RPM)
piRNA Dysregulation in Postmortem Samples
piRNAs are a class of small ncRNA molecules comprising 24–32 nucleotides, which protect genomic DNA mainly in germ cells by association with PIWI proteins, a subfamily of Argonaute proteins [12, 13]. Five piRNAs were dysregulated in sALS samples (Fig. 2B). Among them, three piRNAs were up-regulated (hsa-piR-000578, hsa-piR-020871, and hsa-piR-022184) and two piRNAs were down-regulated (hsa-piR-009294 and hsa-piR-016735), using a false discovery rate (FDR) < 0.05 and p < 0.05 (Table 3).
There are no previous reports of these piRNAs in human brain. The piRNA sequence and target gene sequences were extracted from the PIWI-interacting RNA (piRNA) Database—piRNAdb [14] and predicted target genes for validation from piRBase [15] (Table 4). hsa-piR-33151, which was reported as decreased in serum samples from ALS patients [9], was not detected in our brain samples.
Dysregulation of the five piRNAs was validated using RT-qPCR with two methods, the standard curve method to visualize the overall trend for piRNA perturbation and the △CT method to quantify expression in individual samples. RT-qPCR results confirmed the RNA-Seq analysis and these five piRNAs were significantly altered in sALS samples, in comparison with controls (Fig. 3).
Validation of piRNA expression with RT-qPCR. Dysregulation of five piRNAs was validated using RT-qPCR with two methods, the standard curve method to visualize the overall trend for piRNA perturbation (A) and the △CT method to quantify expression in individual samples (B). Both methods confirm that these five piRNAs were significantly altered in sALS samples in comparison with controls. A Average change in expression of each piRNA in control vs sALS samples. The Wilcoxon signed-rank test analysis was used for statistical calculations. Error bars denote SEMs. B piRNA expression by ΔCT in control vs sALS. The Mann–Whitney test was used for statistical calculations. ns: not significant. *p < 0.05, **p < 0.01
Expression of piRNA Target Genes in Postmortem Samples
The △CT method was used to investigate the change in expression of in silico predicted potential target genes for dysregulated piRNA, according to piRBase and piRNAdb. This computer prediction method has limitations and may indicate false positive targets. Predicted target genes are listed in Tables 4. Most target genes were ribosomal protein pseudogenes, RPL10P7, RPL13AP3, RPL18AP3, and 5S ribosomal RNA pseudogene transcripts, RNA5SP202 and RNA5-8SP6 except TXNRD1 coding gene. Due to increased evidence of ribosomal protein dysregulation in ALS [16], we investigated expression of ribosomal protein coding genes instead of pseudogenes (Fig. 4). There were no notable changes, except for expression of RPL13A (p = 0.053). We also investigated the downstream translation effect on target genes. For this purpose, we checked the expression level of TXRND1. TXRND1 expression was altered in ALS patients; however, the observation was not statistically significant (Supplementary Fig. 1). These data suggest that these piRNAs have regulatory effects beyond in silico-predicted target coding gene regulation.
piRNA target genes are mostly ribosomal protein pseudogenes. The △CT method was used to investigate changes in expression of in silico-predicted target genes for dysregulated piRNA, according to piRBase and piRNAdb (Table 4). Except for TXRND1, target genes included ribosomal protein pseudogenes, RPL10P7, RPL13AP3, RPL18AP3, and 5S ribosomal RNA pseudogene transcripts, RNA5SP202 and RNA5-8SP6. We investigated the coding gene expression instead of pseudogenes due to increased evidence of ribosomal protein dysregulation in ALS. There was no notable change in gene expression except for expression of RPL13A (p = 0.053). This suggests that these piRNAs have regulatory effects beyond regulation of in silico-predicted target coding genes
Dysregulation of PIWI Proteins in Postmortem Samples
piRNAs are produced by PIWI proteins from different genomic loci and function via the PIWI/piRNA complex. Therefore, we investigated expression differences of PIWI proteins in postmortem samples using RT-qPCR and immunoblotting. PIWIL2 and PIWIL3 were not detected in immunoblots in any of the samples (data not shown). RT-qPCR using the △CT method for PIWIL2 and PIWIL3 showed non-significant differences in △CT between sALS and control samples (Fig. 5A). PIWIL1 was upregulated 1.2–1.9-fold in sALS patients (Fig. 5B) compared to the mean of control samples using the standard curve method. △CT for PIWIL1 was reduced in sALS samples, which means an increase in RNA expression (Fig. 5A). The increase of PIWIL1 protein was confirmed by immunoblot (Fig. 5C, D). PIWIL4 was significantly downregulated an average of 0.6-fold in sALS patients (Fig. 5B) compared to control samples, using the standard curve method. △CT for PIWIL4 was increased in sALS samples, indicating a decrease in RNA expression (Fig. 5A). The decrease of PIWIL4 protein was also confirmed by immunoblot (Fig. 5C, D).
PIWI proteins were dysregulated in postmortem sALS samples. A RT-qPCR for PIWIL1, PIWIL2, PIWIL3, PIWIL4 expression of mRNA using the ΔCT method. RT-qPCR using the △CT method for PIWIL1 and PIWIL4 showed significant differences between ALS and control samples, while PIWIL2 and PIWIL3 showed non-significant differences. B RT-qPCR results of postmortem PIWIL1 and PIWIL4 expression using a control pool for the standard curve method. One-tailed t-test and the Wilcoxon test were used for statistical calculations. PIWIL1 was upregulated 1.2–1.9-fold in sALS patients compared to the mean of control samples, by the standard curve method. C Western blot analysis of PIWIL1 and PIWIL4 in postmortem samples. The increase of PIWIL1 protein and the decrease of PIWIL4 protein were confirmed in ALS samples. D Quantitative analysis of PIWIL1 and PIWIL4 in a Western blot normalized to GAPDH. The increase of PIWIL1 protein and the decrease of PIWIL4 protein were statistically confirmed. ns: not significant. *p < 0.05, **p < 0.01
PIWIL1-TDP-43 Colocalization in Postmortem Samples
We examined the expression pattern of PIWIL1 in the lumbar cord (L5) of sALS patients. In control cases, without neurodegenerative diseases, cell nuclei, nuclear membranes, and cytoplasm of anterior horn cells (AHCs) were positive for PIWIL1 (Fig. 6A).
Immunohistochemistry for PIWIL1 in the lumbar spinal cord of control (A) and ALS (B–E) patients. A In control patients, cell nuclei, the nuclear membrane (small arrows), and cell bodies of anterior horn cells (AHCs) are immunopositive for PIWIL1. B. In ALS patients, some remaining AHCs have nuclei positive for PIWIL1 (white arrow), while others have PIWIL1-negative nuclei (black arrow). C–E. In double immunofluorescence staining, PIWIL1 is co-localized with intracytoplasmic inclusions positive for TDP-43 (white arrows) in ALS patients. Scale bar = 20 µm
In the lumbar cord from sALS patients, AHCs were reduced in number and 53% of remaining AHCs were atrophic with PIWIL1-negative nuclei (Fig. 6B). In double immunofluorescence staining, PIWIL1 in cell bodies was partially co-localized with TDP-43-positive cytoplasmic inclusions in 30% of AHCs of sALS patients (Fig. 6C–E and Supplementary Table 2).
Discussion
piRNA Dysregulation in sALS
We detected and validated dysregulation of five piRNAs in postmortem sALS samples. This is the first evidence of piRNA expression changes in brains of ALS patients, as well as the first report of these piRNAs in human brain. RT-qPCR validation of in silico piRNA target genes did not indicate significant alterations in their expression, suggesting that piRNAs have regulatory effects beyond in silico-predicted target coding-gene regulation. Another possible interpretation of these piRNA alterations is that they are a byproduct or a manifestation of dysregulation in a different genomic region.
Generally, piRNAs contribute to silencing of transposable element (TE) expression, chromatin modifications [17], and mRNA localization [18]. The piRNA pathway targets a subset of LINE1 loci that contain full-length retrotransposon insertions [19], and there is growing evidence suggesting that PIWI proteins and piRNAs function mainly as transposable element regulators in the germline. Brains showing increased expression of retrotransposon elements are likely to be a source of altered piRNA production [20]. On the other hand, TDP-43 was reported to suppress transposable elements in ALS [21], and the loss of nuclear TDP-43 is associated with de-condensation of LINE retrotransposons [19]. The fact that all sporadic ALS samples used in this study manifested TDP-43 proteinopathy suggests that TDP-43 does not protect against TE, which may explain the dysregulation of piRNAs in postmortem medulla samples.
PIWI Protein Dysregulation as a Potential Cause of ALS
Our results demonstrate dysregulation in PIWIL1 and PIWIL4 in sALS samples. In humans, four PIWI proteins, PIWIL1-4, are involved in piRNA biogenesis and function. Expression of PIWI proteins differs by tissue and organ [22]. PIWIL1 regulates neuronal polarization and radial migration partly by modulating expression of microtubule-associated proteins (MAPs). MAPs are involved in axon extension by directly binding and stabilizing mRNA of target genes, including MAP1B [23]. Previous observations show the protective effect of PIWIL1 in neuronal development and microtubule migration. Likewise, a recent study using postmortem samples of brains of Alzheimer’s disease patients showed upregulation of PIWIL1 [24]. Together these lines of evidence suggest that altered expression of PIWIL1 can be explained as a compensatory mechanism for axonal dysfunction. On the other hand, PIWIL1 was previously reported to interact physically with TDP-43 in Drosophila [25]. Immunohistochemistry results show PIWIL1 depletion in nuclei of sALS patients compared to controls. Moreover, colocalization of PIWIL1 and TDP-43 was also observed in the cytoplasm of sALS samples, suggesting that increased PIWIL1 and its interaction with TDP-43 in the cytoplasm may contribute to formation of TDP-43 inclusions. This also supports the idea that loss of function in the nucleus or compartmental mislocalization of PIWIL1 in neurons has a drastic impact on neurodegeneration in ALS.
In the current study, PIWIL2 expression was not detected in sALS postmortem samples, supporting the observation by Gasparini et al. [26], that PIWIL2 is expressed in neural stem/progenitor cells, but not in neurons. PIWIL3 also was not detected by immunoblotting in these samples, while PIWIL4 was downregulated by 0.5–0.6-fold in most sALS samples. PIWIL3 has not been reported previously in brain, as it has in ovary, testis, and blood [15]. The role of PIWIL4 in brain is poorly understood, despite recent reports indicating that PIWIL2 and PIWIL4 are associated with autism [27], and that PIWIL4 helps modulate neuronal differentiation from human embryonal carcinoma cells [28]. Disturbance of these tissue specificities of PIWI proteins, or dysfunction of PIWIL4 may also contribute to the disease signature of ALS.
Since PIWIL alterations (PIWL1 and PIWIL4) are observed in the medulla oblongata of ALS samples used in this study, one would expect greater impact on piRNA dysregulation. Technical limitations might explain why only few piRNAs are dysregulated in the same tissue. This could be due to the fact that discovery rate reflects the sequencing depth and the number of useable reads, which may have limited the detection of more target piRNAs. On the other hand, in this study, we have not checked the piRNA population in other areas containing the cell bodies of motor neurons, which might have greater impact.
Overall, our results suggest that dysregulation of the piRNA-PIWI protein axis is associated with pathogenesis of ALS. PIWI protein mislocalization could be an important determinant of TDP-43 accumulation in the cytoplasm. Moreover, dysregulated piRNA and PIWI proteins may be useful as diagnostic biomarkers, as well as gene targets for molecular therapy of ALS.
Conclusions
This study is the first report of the involvement of PIWI/piRNA metabolism in sALS brain tissues. Our results demonstrate significant dysregulation of five piRNAs, as well as PIWI proteins. They indicate that these piRNAs and PIWI proteins are involved in pathogenesis of ALS, and may be useful as specific disease biomarkers. Furthermore, they may eventually serve as targets for development of novel therapeutics, such as nucleic acid medicines or gene therapeutics.
Data Availability
Datasets generated and analyzed during the current study are available in the DNA Data Bank of Japan (DDBJ) under Temporary Submission ID: SSUB016807.
Abbreviations
- ALS:
-
Amyotrophic lateral sclerosis
- fALS:
-
Familial amyotrophic lateral sclerosis
- sALS:
-
Sporadic amyotrophic lateral sclerosis
- ncRNA:
-
Non-coding RNA
- sRNA:
-
Small RNA
- miRNA:
-
Micro RNA
- piRNA:
-
PIWI-interacting RNA
- FDR:
-
False discovery rate
- PIWIL1:
-
PIWI-like RNA-mediated gene silencing 1
- PIWIL2:
-
PIWI-like RNA-mediated gene silencing 2
- PIWIL3:
-
PIWI-like RNA-mediated gene silencing 3
- PIWIL4:
-
PIWI-like RNA-mediated gene silencing 4
- RT-qPCR:
-
Reverse transcription-quantitative PCR
- TDP-43:
-
TAR DNA-binding protein 43
- AHC:
-
Anterior horn cell
- IF:
-
Immunofluorescence
- MAP:
-
Microtubule-associated protein
References
Mejzini R, Flynn LL, Pitout IL et al (2019) ALS genetics, mechanisms, and therapeutics: where are we now? Front Neurosci 13:1310. https://doi.org/10.3389/fnins.2019.01310
Prasad A, Bharathi V, Sivalingam V et al (2019) Molecular mechanisms of TDP-43 misfolding and pathology in amyotrophic lateral sclerosis. Front Mol Neurosci 12:25. https://doi.org/10.3389/fnmol.2019.00025
Wakisaka KT, Imai Y (2019) The dawn of pirna research in various neuronal disorders. Front Biosci Landmark Ed 24:1440–1451
Mao Q, Fan L, Wang X et al (2019) Transcriptome-wide piRNA profiling in human brains for aging genetic factors. Jacobs J Genet 4:014
Brettschneider J, Del Tredici K, Toledo JB et al (2013) Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann Neurol 74:20–38. https://doi.org/10.1002/ana.23937
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal 17:10. https://doi.org/10.14806/ej.17.1.200
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma Oxf Engl 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Kitchen R. exceRpt: The extra-cellular RNA processing toolkit. Includes software to preprocess, align, quantitate, and normalise smallRNA-seq datasets. https://rkitchen.github.io/exceRpt/
Waller R, Wyles M, Heath PR et al (2018) Small RNA sequencing of sporadic amyotrophic lateral sclerosis cerebrospinal fluid reveals differentially expressed miRNAs related to neural and glial activity. Front Neurosci 11:731. https://doi.org/10.3389/fnins.2017.00731
Waller R, Goodall EF, Milo M et al (2017) Serum miRNAs miR-206, 143–3p and 374b–5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiol Aging 55:123–131. https://doi.org/10.1016/j.neurobiolaging.2017.03.027
Freischmidt A, Müller K, Ludolph AC, Weishaupt JH (2013) Systemic dysregulation of TDP-43 binding microRNAs in amyotrophic lateral sclerosis. Acta Neuropathol Commun 1:42. https://doi.org/10.1186/2051-5960-1-42
Girard A, Sachidanandam R, Hannon GJ, Carmell MA (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442:199–202. https://doi.org/10.1038/nature04917
Vagin VV, Sigova A, Li C et al (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science. https://doi.org/10.1126/science.1129333
Piuco R, Galante PAF. piRNAdb. https://www.pirnadb.org/index
Wang J, Zhang P, Lu Y et al (2019) piRBase: a comprehensive database of piRNA sequences. Nucleic Acids Res 47:D175–D180. https://doi.org/10.1093/nar/gky1043
Nagano S, Jinno J, Abdelhamid RF et al (2020) TDP-43 transports ribosomal protein mRNA to regulate axonal local translation in neuronal axons. Acta Neuropathol (Berl) 140:695–713. https://doi.org/10.1007/s00401-020-02205-y
Yamanaka S, Siomi MC, Siomi H (2014) piRNA clusters and open chromatin structure. Mob DNA 5:22. https://doi.org/10.1186/1759-8753-5-22
Dufourt J, Bontonou G, Chartier A et al (2017) piRNAs and Aubergine cooperate with Wispy poly(A) polymerase to stabilize mRNAs in the germ plasm. Nat Commun 8:1305. https://doi.org/10.1038/s41467-017-01431-5
Pezic D, Manakov SA, Sachidanandam R, Aravin AA (2014) piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev 28:1410–1428. https://doi.org/10.1101/gad.240895.114
Nandi S, Chandramohan D, Fioriti L et al (2016) Roles for small noncoding RNAs in silencing of retrotransposons in the mammalian brain. Proc Natl Acad Sci 113:12697–12702. https://doi.org/10.1073/pnas.1609287113
Tam OH, Ostrow LW, Gale Hammell M (2019) Diseases of the nERVous system: retrotransposon activity in neurodegenerative disease. Mob DNA 10:32. https://doi.org/10.1186/s13100-019-0176-1
Rojas-Ríos P, Simonelig M (2018) piRNAs and PIWI proteins: regulators of gene expression in development and stem cells. Development 145:dev161786. https://doi.org/10.1242/dev.161786
Zhao P, Yao M, Chang S et al (2015) Novel function of PIWIL1 in neuronal polarization and migration via regulation of microtubule-associated proteins. Mol Brain 8:39. https://doi.org/10.1186/s13041-015-0131-0
Frost B, Hemberg M, Lewis J, Feany MB (2014) Tau promotes neurodegeneration through global chromatin relaxation. Nat Neurosci 17:357–366. https://doi.org/10.1038/nn.3639
Godena VK, Romano G, Romano M et al (2011) TDP-43 Regulates Drosophila neuromuscular junctions growth by modulating futsch/MAP1B levels and synaptic microtubules organization. PLoS ONE 6:e17808. https://doi.org/10.1371/journal.pone.0017808
Gasperini C, Pelizzoli R, Van AL et al (2020) The piRNA pathway sustains adult neurogenesis by repressing protein synthesis. https://doi.org/10.1101/2020.09.15.297739
Iossifov I, O’Roak BJ, Sanders SJ et al (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature 515:216–221. https://doi.org/10.1038/nature13908
Subhramanyam CS, Cao Q, Wang C et al (2020) Role of PIWI-like 4 in modulating neuronal differentiation from human embryonal carcinoma cells. RNA Biol 17:1613–1624. https://doi.org/10.1080/15476286.2020.1757896
Acknowledgements
This study was supported in part by Genome Information Research Center, Osaka University.
Funding
This work was funded by Grants-in-Aid for Scientific Research (C) (22590932, 25461302 and 16K09690 to SN), a Grant-in-Aid for Scientific Research on Innovative Areas (16H06277 to SN), AMED (JP20lm0203007 and JP20ek0109320 to SN, JP18dm0107103 to YS), grants from the Japan Foundation for Neuroscience and Mental Health and Strategic Research Program for Brain Sciences (to SN). The funding bodies did not contribute to the design of the study, to collection, analysis, or interpretation of data, or to the writing of the manuscript.
Author information
Authors and Affiliations
Contributions
RFA, SN, KO, GB, KI, KB, YN, YO, and HM developed and organized the study. RFA, SN, KO, GB, ET, YY and JJ performed experiments. YS and SM collected and evaluated human samples. RFA and SN wrote the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Collection of autopsied human brain tissues and their use in this study were approved by the Ethics Committees of Osaka University Graduate School of Medicine, Tokyo Metropolitan Geriatric Hospital and Institute of Gerontology (TMGHIG) and the National Center of Neurology and Psychiatry (NCNP). Written informed consent was obtained from patient families. Experimental procedures involving human subjects were conducted in conformance with principles of the Declaration of Osaka University.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abdelhamid, R.F., Ogawa, K., Beck, G. et al. piRNA/PIWI Protein Complex as a Potential Biomarker in Sporadic Amyotrophic Lateral Sclerosis. Mol Neurobiol 59, 1693–1705 (2022). https://doi.org/10.1007/s12035-021-02686-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-021-02686-2