Abstract
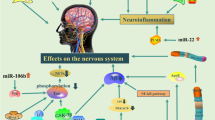
LncRNAs have emerged as important regulatory molecules in biological processes. They serve as regulators of gene expression pathways through interactions with proteins, RNA, and DNA. LncRNA expression is altered in several diseases of the central nervous system (CNS), such as neurodegenerative disorders, stroke, trauma, and infection. More recently, it has become clear that lncRNAs contribute to regulating both pro-inflammatory and anti-inflammatory pathways in the CNS. In this review, we discuss the molecular pathways involved in the expression of lncRNAs, their role and mechanism of action during gene regulation, cellular functions, and use of lncRNAs as therapeutic targets during neuroinflammation in CNS disorders.


Similar content being viewed by others
Data Availability
Not applicable
Abbreviations
- AD :
-
Alzheimer’s disease
- ALS:
-
amyotrophic lateral sclerosis
- ANRIL:
-
antisense non-coding RNA in the INK4 locus
- APP :
-
amyloid precursor protein
- Aβ:
-
amyloid beta
- BACE1:
-
β-site amyloid precursor protein cleaving enzyme 1
- BBB:
-
blood-brain barrier
- BDNF:
-
brain-derived neurotrophic factor
- BMECs:
-
brain microvascular endothelial cells
- C2dat1 :
-
CAMK2D-associated transcript 1
- CCAT1:
-
colon cancer–associated transcript-1
- CCL2:
-
C-C motif chemokine ligand 2
- ceRNAs:
-
competing endogenous RNAs
- CNS :
-
central nervous system
- CRNDE:
-
colorectal neoplasia differentially expressed
- DGCR5:
-
DiGeorge syndrome critical region gene 5
- DILC:
-
LncRNA downregulated in liver cancer stem cells
- DM:
-
diabetes mellitus
- DRG:
-
dorsal root ganglion
- EAE:
-
experimental autoimmune encephalomyelitis
- EBI:
-
early brain injury
- EZH2:
-
enhancer of zester homolog 2
- FAL1:
-
focally amplified lncRNA on chromosome 1
- FIRRE:
-
functional intergenic repeating RNA element
- FosDT:
-
Fos downstream transcript
- FUS:
-
fused in sarcoma
- GAS5:
-
growth arrest-specific 5
- GBM:
-
glioblastoma multiforme
- GBP9:
-
LncRNA guanylate binding protein-9
- GFAP:
-
glial fibrillary acidic protein
- GO:
-
gene ontology
- HIV-1:
-
human immunodeficiency virus
- HMGB-1:
-
high mobility group box-1
- HnRNP-A2/B1:
-
heterogeneous nuclear ribonucleoprotein A/B and A2/B1
- I/R:
-
ischemia and reperfusion
- ICH:
-
intracerebral hemorrhage
- IFN-γ:
-
interferon-gamma
- IL-1β :
-
interleukin-1β
- IL-6 :
-
interleukin-6
- iNOS:
-
inducible nitric oxide synthases
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- lincRNAs:
-
long intergenic non-coding RNAs
- lncRNAs:
-
long non-coding RNAs
- LPS:
-
lipopolysaccharide
- Maclpil:
-
macrophage contained LCP1-related pro-inflammatory
- MALAT1:
-
metastasis-associated lung adenocarcinoma transcript 1
- MAPK :
-
mitogen-activated protein kinase
- MCAO:
-
middle cerebral artery occlusion mouse
- MCP-1:
-
monocyte chemoattractant protein-1
- miRNAs:
-
microRNAs
- MPTP:
-
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- MS :
-
multiple sclerosis
- Mtss1:
-
metastasis suppressor-1
- ncRNAs:
-
non-coding RNAs
- NDM29:
-
neuroblastoma differentiation marker 29
- NEAT1:
-
nuclear paraspeckle assembly transcript 1
- NF-κB:
-
nuclear factor-k-gene binding
- NKILA:
-
NF-κB interacting lncRNA
- NLRP3:
-
Nod-like receptor protein 3
- NO:
-
nitric oxide
- NRON:
-
non-coding repressor of nuclear factor of activated T cells
- OGD/R:
-
oxygen glucose deprivation/reoxygenation
- PCAI:
-
protect cells from apoptosis and inflammation
- PD:
-
Parkinson’s disease
- PI3K :
-
phosphoinositide 3-kinase
- PKC:
-
protein kinase C
- PRC2:
-
polycomb repressive complex 2
- RGCs:
-
retinal ganglion cells
- RMST:
-
rhabdomyosarcoma 2–associated transcript
- ROS:
-
reactive oxygen species
- SAE:
-
sepsis-associated encephalopathy
- SAH:
-
subarachnoid hemorrhage
- SCI:
-
spinal cord injury
- siRNAs:
-
small interfering RNAs
- SNL:
-
spinal nerve ligation
- SOCS3:
-
suppressor of cytokine signaling 3
- SOD1:
-
superoxide dismutase 1
- STAT6:
-
signal transduction and activators of transcription 6
- TAK1:
-
TGF-β-activated kinase 1
- TARDBP:
-
transactive response DNA binding protein
- TBI:
-
traumatic brain injury
- TGF-β:
-
transforming growth factor
- TLE:
-
temporal lobe epilepsy
- TNF-α :
-
tumor necrosis factor-α
- TUG1:
-
taurine-upregulated gene 1
- VEGF:
-
vascular endothelial growth factor
- XIST:
-
X inactive specific transcript
References
Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J et al (2012) Landscape of transcription in human cells. Nature 489:101–108. https://doi.org/10.1038/nature11233
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20:675–691. https://doi.org/10.1038/s41576-019-0158-7
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. https://doi.org/10.1038/nature11928
Bartel DP (2018) Metazoan microRNAs. Cell 173:20–51. https://doi.org/10.1016/j.cell.2018.03.006
Czech B, Munafò M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, Hannon GJ (2018) piRNA-guided genome defense: from biogenesis to silencing. Annu Rev Genet 52:131–157. https://doi.org/10.1146/annurev-genet-120417-031441
Rajappa A, Banerjee S, Sharma V, Khandelia P (2020) Circular RNAs: emerging role in cancer diagnostics and therapeutics. Front Mol Biosci 7:577938. https://doi.org/10.3389/fmolb.2020.577938
Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166. https://doi.org/10.1146/annurev-biochem-051410-092902
Bonasio R, Shiekhattar R (2014) Regulation of transcription by long noncoding RNAs. Annu Rev Genet 48:433–455. https://doi.org/10.1146/annurev-genet-120213-092323
Fernandes J, Acuña S, Aoki J, Floeter-Winter L, Muxel S (2019) Long non-coding RNAs in the regulation of gene expression: physiology and disease. Non-Coding RNA 5:17. https://doi.org/10.3390/ncrna5010017
Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR et al (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47:199–208. https://doi.org/10.1038/ng.3192
Zhao Y, Li H, Fang S, Kang Y, wu W, Hao Y, Li Z, Bu D et al (2016) NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res 44:D203–D208. https://doi.org/10.1093/nar/gkv1252
St Laurent G, Wahlestedt C, Kapranov P (2015) The landscape of long noncoding RNA classification. Trends Genet TIG 31:239–251. https://doi.org/10.1016/j.tig.2015.03.007
Gil N, Ulitsky I (2020) Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet 21:102–117. https://doi.org/10.1038/s41576-019-0184-5
Rinn JL, Chang HY (2020) Long noncoding RNAs: molecular modalities to organismal functions. Annu Rev Biochem 89:283–308. https://doi.org/10.1146/annurev-biochem-062917-012708
Kopp F, Mendell JT (2018) Functional classification and experimental dissection of long noncoding RNAs. Cell 172:393–407. https://doi.org/10.1016/j.cell.2018.01.011
Chen S, Dong Z, Cheng M, Zhao Y, Wang M, Sai N, Wang X, Liu H et al (2017) Homocysteine exaggerates microglia activation and neuroinflammation through microglia localized STAT3 overactivation following ischemic stroke. J Neuroinflammation 14:187. https://doi.org/10.1186/s12974-017-0963-x
Xanthos DN, Sandkühler J (2014) Neurogenic neuroinflammation: inflammatory CNS reactions in response to neuronal activity. Nat Rev Neurosci 15:43–53. https://doi.org/10.1038/nrn3617
Lyman M, Lloyd DG, Ji X, Vizcaychipi MP, Ma D (2014) Neuroinflammation: the role and consequences. Neurosci Res 79:1–12. https://doi.org/10.1016/j.neures.2013.10.004
Shabab T, Khanabdali R, Moghadamtousi SZ, Kadir HA, Mohan G (2017) Neuroinflammation pathways: a general review. Int J Neurosci 127:624–633. https://doi.org/10.1080/00207454.2016.1212854
Das S, Basu A (2008) Inflammation: a new candidate in modulating adult neurogenesis. J Neurosci Res 86:1199–1208. https://doi.org/10.1002/jnr.21585
Tandon PN (2017) The enigma of neuroinflammation. Neurol India 65:703–705. https://doi.org/10.4103/neuroindia.NI_517_17
Brown CM, Mulcahey TA, Filipek NC, Wise PM (2010) Production of proinflammatory cytokines and chemokines during neuroinflammation: novel roles for estrogen receptors α and β. Endocrinology 151:4916–4925. https://doi.org/10.1210/en.2010-0371
Mittal M, Siddiqui MR, Tran K, Reddy SP, Malik AB (2014) Reactive oxygen species in inflammation and tissue injury. Antioxid Redox Signal 20:1126–1167. https://doi.org/10.1089/ars.2012.5149
Ransohoff RM (2016) How neuroinflammation contributes to neurodegeneration. Science 353:777–783. https://doi.org/10.1126/science.aag2590
Ransohoff RM, Perry VH (2009) Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol 27:119–145. https://doi.org/10.1146/annurev.immunol.021908.132528
Hanisch U-K, Kettenmann H (2007) Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci 10:1387–1394. https://doi.org/10.1038/nn1997
DiSabato DJ, Quan N, Godbout JP (2016) Neuroinflammation: the devil is in the details. J Neurochem 139(Suppl 2):136–153. https://doi.org/10.1111/jnc.13607
Kempuraj D, Thangavel R, Natteru PA et al (2016) Neuroinflammation induces neurodegeneration. J Neurol Neurosurg Spine 1:1003
Guo Y, Hong W, Wang X, Zhang P, Körner H, Tu J, Wei W (2019) MicroRNAs in microglia: how do microRNAs affect activation, inflammation, polarization of microglia and mediate the interaction between microglia and glioma? Front Mol Neurosci 12:125. https://doi.org/10.3389/fnmol.2019.00125
Slota JA, Booth SA (2019) MicroRNAs in neuroinflammation: implications in disease pathogenesis, biomarker discovery and therapeutic applications. Non-Coding RNA 5:35. https://doi.org/10.3390/ncrna5020035
Li R, Zhu H, Luo Y (2016) understanding the functions of long non-coding RNAs through their higher-order structures. Int J Mol Sci 17. https://doi.org/10.3390/ijms17050702
Shree B, Sharma V (2018) Linc'ing' RNA to DNA repair (2018). Proc Indian Natn Sci Acad 84(2):521–529. https://doi.org/10.6943/ptinsa/2018/49332
Ulitsky I, Bartel DP (2013) lincRNAs: genomics, evolution, and mechanisms. Cell 154:26–46. https://doi.org/10.1016/j.cell.2013.06.020
Salmena L, Poliseno L, Tay Y et al (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–358. https://doi.org/10.1016/j.cell.2011.07.014
Cerase A, Pintacuda G, Tattermusch A, Avner P (2015) Xist localization and function: new insights from multiple levels. Genome Biol 16:166. https://doi.org/10.1186/s13059-015-0733-y
Rinn JL, Kertesz M, Wang JK et al (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129:1311–1323. https://doi.org/10.1016/j.cell.2007.05.022
Carpenter S, Fitzgerald KA (2015) Transcription of inflammatory genes: long noncoding RNA and beyond. J Interferon Cytokine Res Off J Int Soc Interferon Cytokine Res 35:79–88. https://doi.org/10.1089/jir.2014.0120
Peng X, Gralinski L, Armour CD et al (2010) Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio:1. https://doi.org/10.1128/mBio.00206-10
Carpenter S, Aiello D, Atianand MK et al (2013) A long noncoding RNA mediates both activation and repression of immune response genes. Science 341:789–792. https://doi.org/10.1126/science.1240925
Carpenter S, Fitzgerald KA (2018) Cytokines and Long noncoding RNAs. Cold Spring Harb Perspect Biol 10:a028589. https://doi.org/10.1101/cshperspect.a028589
Wen Y, Yu Y, Fu X (2017) LncRNA Gm4419 contributes to OGD/R injury of cerebral microglial cells via IκB phosphorylation and NF-κB activation. Biochem Biophys Res Commun 487:923–929. https://doi.org/10.1016/j.bbrc.2017.05.005
Sun D, Yu Z, Fang X et al (2017) LncRNA GAS5 inhibits microglial M2 polarization and exacerbates demyelination. EMBO Rep 18:1801–1816. https://doi.org/10.15252/embr.201643668
Xue Z, Zhang Z, Liu H et al (2019) lincRNA-Cox2 regulates NLRP3 inflammasome and autophagy mediated neuroinflammation. Cell Death Differ 26:130–145. https://doi.org/10.1038/s41418-018-0105-8
Ye Y, He X, Lu F et al (2018) A lincRNA-p21/miR-181 family feedback loop regulates microglial activation during systemic LPS- and MPTP- induced neuroinflammation. Cell Death Dis 9:803. https://doi.org/10.1038/s41419-018-0821-5
Gelders G, Baekelandt V, Van der Perren A (2018) linking neuroinflammation and neurodegeneration in parkinson’s disease. J Immunol Res 2018:1–12. https://doi.org/10.1155/2018/4784268
Guzman-Martinez L, Maccioni RB, Andrade V et al (2019) Neuroinflammation as a Common Feature of Neurodegenerative Disorders. Front Pharmacol 10:1008. https://doi.org/10.3389/fphar.2019.01008
Tian L, Ma L, Kaarela T, Li Z (2012) Neuroimmune crosstalk in the central nervous system and its significance for neurological diseases. J Neuroinflammation 9:155. https://doi.org/10.1186/1742-2094-9-155
Liberman AC, Trias E, da Silva CL et al (2018) Neuroimmune and Inflammatory Signals in Complex Disorders of the Central Nervous System. Neuroimmunomodulation 25:246–270. https://doi.org/10.1159/000494761
Ramesh G, MacLean AG, Philipp MT (2013) Cytokines and chemokines at the crossroads of neuroinflammation, neurodegeneration, and neuropathic pain. Mediators Inflamm 2013:480739. https://doi.org/10.1155/2013/480739
Leyns CEG, Holtzman DM (2017) Glial contributions to neurodegeneration in tauopathies. Mol Neurodegener 12:50. https://doi.org/10.1186/s13024-017-0192-x
Faghihi MA, Modarresi F, Khalil AM et al (2008) Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 14:723–730. https://doi.org/10.1038/nm1784
Faghihi MA, Zhang M, Huang J et al (2010) Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol 11:R56. https://doi.org/10.1186/gb-2010-11-5-r56
Zagrebelsky M, Korte M (2014) Form follows function: BDNF and its involvement in sculpting the function and structure of synapses. Neuropharmacology 76 Pt C:628–638. https://doi.org/10.1016/j.neuropharm.2013.05.029
Lima Giacobbo B, Doorduin J, Klein HC et al (2019) Brain-derived neurotrophic factor in brain disorders: focus on neuroinflammation. Mol Neurobiol 56:3295–3312. https://doi.org/10.1007/s12035-018-1283-6
O’Bryant SE, Hobson V, Hall JR et al (2009) Brain-derived neurotrophic factor levels in Alzheimer’s disease. J Alzheimers Dis JAD 17:337–341. https://doi.org/10.3233/JAD-2009-1051
Modarresi F, Faghihi MA, Lopez-Toledano MA et al (2012) Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol 30:453–459. https://doi.org/10.1038/nbt.2158
Massone S, Vassallo I, Fiorino G et al (2011) 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis 41:308–317. https://doi.org/10.1016/j.nbd.2010.09.019
Massone S, Ciarlo E, Vella S et al (2012) NDM29, a RNA polymerase III-dependent non coding RNA, promotes amyloidogenic processing of APP and amyloid β secretion. Biochim Biophys Acta 1823:1170–1177. https://doi.org/10.1016/j.bbamcr.2012.05.001
Zhou Y, Xu Z, Yu Y et al (2019) Comprehensive analysis of the lncRNA-associated ceRNA network identifies neuroinflammation biomarkers for Alzheimer’s disease. Mol Omics 15:459–469. https://doi.org/10.1039/C9MO00129H
Ma P, Li Y, Zhang W, Fang F, Sun J, Liu M, Li K, Dong L (2019) Long Non-coding RNA MALAT1 inhibits neuron apoptosis and neuroinflammation while stimulates neurite outgrowth and its correlation with MiR-125b mediates PTGS2, CDK5 and FOXQ1 in alzheimer's disease. Current Alzheimer Research 16(7):596–612. https://doi.org/10.2174/1567205016666190725130134
Yi J, Chen B, Yao X et al (2019) Upregulation of the lncRNA MEG3 improves cognitive impairment, alleviates neuronal damage, and inhibits activation of astrocytes in hippocampus tissues in Alzheimer’s disease through inactivating the PI3K/Akt signaling pathway. J Cell Biochem 120:18053–18065. https://doi.org/10.1002/jcb.29108
Zhou B, Li L, Qiu X et al (2020) Long non-coding RNA ANRIL knockdown suppresses apoptosis and pro-inflammatory cytokines while enhancing neurite outgrowth via binding microRNA-125a in a cellular model of Alzheimer’s disease. Mol Med Rep 22:1489–1497. https://doi.org/10.3892/mmr.2020.11203
Tang L, Liu L, Li G et al (2019) Expression Profiles of Long Noncoding RNAs in Intranasal LPS-Mediated Alzheimer’s Disease Model in Mice. BioMed Res Int 2019:1–14. https://doi.org/10.1155/2019/9642589
Wang Q, Liu Y, Zhou J (2015) Neuroinflammation in Parkinson’s disease and its potential as therapeutic target. Transl Neurodegener 4:19. https://doi.org/10.1186/s40035-015-0042-0
Chen Y, Lian Y, Ma Y et al (2018) LncRNA SNHG1 promotes α-synuclein aggregation and toxicity by targeting miR-15b-5p to activate SIAH1 in human neuroblastoma SH-SY5Y cells. NeuroToxicology 68:212–221. https://doi.org/10.1016/j.neuro.2017.12.001
Cao B, Wang T, Qu Q et al (2018) Long Noncoding RNA SNHG1 Promotes Neuroinflammation in Parkinson’s Disease via Regulating miR-7/NLRP3 Pathway. Neuroscience 388:118–127. https://doi.org/10.1016/j.neuroscience.2018.07.019
Xu W, Zhang L, Geng Y et al (2020) Long noncoding RNA GAS5 promotes microglial inflammatory response in Parkinson’s disease by regulating NLRP3 pathway through sponging miR-223-3p. Int Immunopharmacol 85:106614. https://doi.org/10.1016/j.intimp.2020.106614
Yu S, Liu X, Yu D, et al (2020) Downregulation of long non-coding RNA SNHG7 protects against inflammation and apoptosis in Parkinson’s disease model by targeting miR-425-5p/TRAF5/NF-κB axis. In Review
Cai L, Tu L, Li T et al (2019) Downregulation of lncRNA UCA1 ameliorates the damage of dopaminergic neurons, reduces oxidative stress and inflammation in Parkinson’s disease through the inhibition of the PI3K/Akt signaling pathway. Int Immunopharmacol 75:105734. https://doi.org/10.1016/j.intimp.2019.105734
Yuan D, Wang Q, Ding N et al (2019) LncRNA MALAT1 aggravates MPP-induced neuronal injury by regulating miR-212 in SH-SY5Y cells. RSC Adv 9:690–698. https://doi.org/10.1039/C8RA09260E
Fan Y, Li J, Yang Q et al (2019) Dysregulated Long Non-coding RNAs in Parkinson’s Disease Contribute to the Apoptosis of Human Neuroblastoma Cells. Front Neurosci 13:1320. https://doi.org/10.3389/fnins.2019.01320
Cai L-J, Tu L, Huang X-M et al (2020) LncRNA MALAT1 facilitates inflammasome activation via epigenetic suppression of Nrf2 in Parkinson’s disease. Mol Brain 13:130. https://doi.org/10.1186/s13041-020-00656-8
Liu R, Li F, Zhao W (2020) Long noncoding RNA NEAT1 knockdown inhibits MPP+-induced apoptosis, in?ammation and cytotoxicity in SK-N-SH cells by regulating miR-212-5p/RAB3IP axis. Neurosci Lett 731:135060. https://doi.org/10.1016/j.neulet.2020.135060
Han Y, Kang C, Kang M et al (2019) Long non-coding RNA Mirt2 prevents TNF-α-triggered inflammation via the repression of microRNA-101. Int Immunopharmacol 76:105878. https://doi.org/10.1016/j.intimp.2019.105878
Wootla B, Eriguchi M, Rodriguez M (2012) Is multiple sclerosis an autoimmune disease? Autoimmune Dis 2012:969657. https://doi.org/10.1155/2012/969657
Franco R, Fernández-Suárez D (2015) Alternatively activated microglia and macrophages in the central nervous system. Prog Neurobiol 131:65–86. https://doi.org/10.1016/j.pneurobio.2015.05.003
Yue P, Jing L, Zhao X et al (2019) Down-regulation of taurine-up-regulated gene 1 attenuates inflammation by sponging miR-9-5p via targeting NF-κB1/p50 in multiple sclerosis. Life Sci 233:116731. https://doi.org/10.1016/j.lfs.2019.116731
Masoumi F, Ghorbani S, Talebi F et al (2019) Malat1 long noncoding RNA regulates inflammation and leukocyte differentiation in experimental autoimmune encephalomyelitis. J Neuroimmunol 328:50–59. https://doi.org/10.1016/j.jneuroim.2018.11.013
Fenoglio C, Oldoni E, Serpente M et al (2018) LncRNAs expression profile in peripheral blood mononuclear cells from multiple sclerosis patients. J Neuroimmunol 324:129–135. https://doi.org/10.1016/j.jneuroim.2018.08.008
Liu X, Zhang Q, Wang W et al (2018) Analysis of long noncoding RNA and mRNA expression profiles in IL-9-activated astrocytes and EAE mice. Cell Physiol Biochem 45:1986–1998. https://doi.org/10.1159/000487975
Schirmer L, Velmeshev D, Holmqvist S et al (2019) Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature 573:75–82. https://doi.org/10.1038/s41586-019-1404-z
Wang L, Gutmann DH, Roos RP (2011) Astrocyte loss of mutant SOD1 delays ALS disease onset and progression in G85R transgenic mice. Hum Mol Genet 20:286–293. https://doi.org/10.1093/hmg/ddq463
Corcia P, Tauber C, Vercoullie J et al (2012) Molecular imaging of microglial activation in amyotrophic lateral sclerosis. PLoS ONE 7:e52941. https://doi.org/10.1371/journal.pone.0052941
Brites D, Vaz AR (2014) Microglia centered pathogenesis in ALS: insights in cell interconnectivity. Front Cell Neurosci 8:117. https://doi.org/10.3389/fncel.2014.00117
Gagliardi S, Zucca S, Pandini C et al (2018) Long non-coding and coding RNAs characterization in peripheral blood mononuclear cells and spinal cord from amyotrophic lateral sclerosis patients. Sci Rep 8:2378. https://doi.org/10.1038/s41598-018-20679-5
Bede P, Bokde ALW, Byrne S et al (2012) Spinal cord markers in ALS: diagnostic and biomarker considerations. Amyotroph Lateral Scler Off Publ World Fed Neurol Res Group Mot Neuron Dis 13:407–415. https://doi.org/10.3109/17482968.2011.649760
Zucca S, Gagliardi S, Pandini C et al (2019) RNA-Seq profiling in peripheral blood mononuclear cells of amyotrophic lateral sclerosis patients and controls. Sci Data 6:190006. https://doi.org/10.1038/sdata.2019.6
Bright F, Werry EL, Dobson-Stone C et al (2019) Neuroinflammation in frontotemporal dementia. Nat Rev Neurol 15:540–555. https://doi.org/10.1038/s41582-019-0231-z
Trageser KJ, Smith C, Herman FJ et al (2019) Mechanisms of immune activation by c9orf72-expansions in amyotrophic lateral sclerosis and frontotemporal dementia. Front Neurosci 13:1298. https://doi.org/10.3389/fnins.2019.01298
Galimberti D, Bonsi R, Fenoglio C et al (2015) Inflammatory molecules in frontotemporal dementia: Cerebrospinal fluid signature of progranulin mutation carriers. Brain Behav Immun 49:182–187. https://doi.org/10.1016/j.bbi.2015.05.006
Galimberti D, Scarpini E (2012) Genetics of frontotemporal lobar degeneration. Front Neurol 3. https://doi.org/10.3389/fneur.2012.00052
Lall D, Baloh RH (2017) Microglia and C9orf72 in neuroinflammation and ALS and frontotemporal dementia. J Clin Invest 127:3250–3258. https://doi.org/10.1172/JCI90607
DeJesus-Hernandez M, Mackenzie IR, Boeve BF et al (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 Causes chromosome 9p-linked FTD and ALS. Neuron 72:245–256. https://doi.org/10.1016/j.neuron.2011.09.011
Renton AE, Majounie E, Waite A et al (2011) A hexanucleotide repeat expansion in C9ORF72 Is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72:257–268. https://doi.org/10.1016/j.neuron.2011.09.010
Bevan-Jones WR, Cope TE, Jones PS et al (2020) Neuroinflammation and protein aggregation co-localize across the frontotemporal dementia spectrum. Brain 143:1010–1026. https://doi.org/10.1093/brain/awaa033
Tollervey JR, Curk T, Rogelj B et al (2011) Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14:452–458. https://doi.org/10.1038/nn.2778
Nishimoto Y, Nakagawa S, Hirose T et al (2013) The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol Brain 6:31. https://doi.org/10.1186/1756-6606-6-31
Lagier-Tourenne C, Polymenidou M, Hutt KR et al (2012) Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long premRNAs. Nat Neurosci 15:1488–1497. https://doi.org/10.1038/nn.3230
Zhang L, Wang H (2019) Long non-coding RNA in CNS injuries: a new target for therapeutic intervention. Mol Ther - Nucleic Acids 17:754–766. https://doi.org/10.1016/j.omtn.2019.07.013
Jayaraj RL, Azimullah S, Beiram R et al (2019) Neuroinflammation: friend and foe for ischemic stroke. J Neuroinflammation 16:142. https://doi.org/10.1186/s12974-019-1516-2
Dykstra-Aiello C, Jickling GC, Ander BP et al (2016) Altered expression of long noncoding rnas in blood after ischemic stroke and proximity to putative stroke risk loci. Stroke 47:2896–2903. https://doi.org/10.1161/STROKEAHA.116.013869
Wang S-W, Liu Z, Shi Z-S (2018) Non-Ccoding RNA in acute ischemic stroke: mechanisms, biomarkers and therapeutic targets. Cell Transplant 27:1763–1777. https://doi.org/10.1177/0963689718806818
Bao M-H, Szeto V, Yang BB et al (2018) Long non-coding RNAs in ischemic stroke. Cell Death Dis 9:281. https://doi.org/10.1038/s41419-018-0282-x
Wang J, Cao B, Han D et al (2017) Long non-coding RNA H19 induces cerebral ischemia reperfusion injury via activation of autophagy. Aging Dis 8:71–84. https://doi.org/10.14336/AD.2016.0530
Wang J, Zhao H, Fan Z et al (2017) Long noncoding RNA H19 promotes neuroinflammation in ischemic stroke by driving histone deacetylase 1-dependent M1 microglial polarization. Stroke 48:2211–2221. https://doi.org/10.1161/STROKEAHA.117.017387
Han C-L, Ge M, Liu Y-P et al (2018) LncRNA H19 contributes to hippocampal glial cell activation via JAK/STAT signaling in a rat model of temporal lobe epilepsy. J Neuroinflammation 15:103. https://doi.org/10.1186/s12974-018-1139-z
Zhang X, Tang X, Liu K et al (2017) Long noncoding RNA Malat1 regulates cerebrovascular pathologies in ischemic stroke. J Neurosci Off J Soc Neurosci 37:1797–1806. https://doi.org/10.1523/JNEUROSCI.3389-16.2017
Zang Y, Zhou X, Wang Q et al (2018) LncRNA FIRRE/NF-kB feedback loop contributes to OGD/R injury of cerebral microglial cells. Biochem Biophys Res Commun 501:131–138. https://doi.org/10.1016/j.bbrc.2018.04.194
Wang Y, Luo Y, Yao Y et al (2020) Silencing the lncRNA Maclpil in pro-inflammatory macrophages attenuates acute experimental ischemic stroke via LCP1 in mice. J Cereb Blood Flow Metab Off J Int Soc Cereb Blood Flow Metab 40:747–759. https://doi.org/10.1177/0271678X19836118
Chen J-X, Wang Y-P, Zhang X et al (2020) lncRNA Mtss1 promotes inflammatory responses and secondary brain injury after intracerebral hemorrhage by targeting miR-709 in mice. Brain Res Bull 162:20–29. https://doi.org/10.1016/j.brainresbull.2020.04.017
Mehta SL, Kim T, Vemuganti R (2015) Long noncoding RNA FosDT Promotes ischemic brain injury by interacting with REST-associated chromatin-modifying proteins. J Neurosci Off J Soc Neurosci 35:16443–16449. https://doi.org/10.1523/JNEUROSCI.2943-15.2015
Peng J, Wu Y, Tian X et al (2017) High-throughput sequencing and Co-expression network analysis of lncRNAs and mRNAs in early brain injury following experimental subarachnoid haemorrhage. Sci Rep 7:46577. https://doi.org/10.1038/srep46577
Long F-Q, Su Q-J, Zhou J-X et al (2018) LncRNA SNHG12 ameliorates brain microvascular endothelial cell injury by targeting miR-199a. Neural Regen Res 13:1919–1926. https://doi.org/10.4103/1673-5374.238717
Zhang J, Dong B, Hao J et al (2019) LncRNA Snhg3 contributes to dysfunction of cerebral microvascular cells in intracerebral hemorrhage rats by activating the TWEAK/Fn14/STAT3 pathway. Life Sci 237:116929. https://doi.org/10.1016/j.lfs.2019.116929
Zhong Y, Yu C, Qin W (2019) LncRNA SNHG14 promotes inflammatory response induced by cerebral ischemia/reperfusion injury through regulating miR-136-5p /ROCK1. Cancer Gene Ther 26:234–247. https://doi.org/10.1038/s41417-018-0067-5
Xu Q, Deng F, Xing Z et al (2016) Long non-coding RNA C2dat1 regulates CaMKIIδ expression to promote neuronal survival through the NF-κB signaling pathway following cerebral ischemia. Cell Death Dis 7:e2173. https://doi.org/10.1038/cddis.2016.57
Deng Y, Chen D, Wang L, et al (2019) Silencing of Long Noncoding RNA Nespas Aggravates Microglial Cell Death and Neuroinflammation in Ischemic Stroke. Stroke 50:1850–1858. https://doi.org/10.1161/STROKEAHA.118.023376
Wang H, Liao S, Li H et al (2019) Long non-coding RNA TUG1 Sponges Mir-145a-5p to regulate microglial polarization after oxygen-glucose deprivation. Front Mol Neurosci 12:215. https://doi.org/10.3389/fnmol.2019.00215
Cao D-W, Liu M-M, Duan R et al (2020) The lncRNA Malat1 functions as a ceRNA to contribute to berberine-mediated inhibition of HMGB1 by sponging miR-181c-5p in poststroke inflammation. Acta Pharmacol Sin 41:22–33. https://doi.org/10.1038/s41401-019-0284-y
Sun X, Wang Z, Wu Q et al (2019) LncRNA RMST activates TAK1-mediated NF-κB signaling and promotes activation of microglial cells via competitively binding with hnRNPK. IUBMB Life 71:1785–1793. https://doi.org/10.1002/iub.2125
Li P, Li Y, Dai Y et al (2020) The LncRNA H19/miR-1-3p/CCL2 axis modulates lipopolysaccharide (LPS) stimulation-induced normal human astrocyte proliferation and activation. Cytokine 131:155106. https://doi.org/10.1016/j.cyto.2020.155106
Wang M, Jiang Y-M, Xia L-Y et al (2018) LncRNA NKILA upregulation mediates oxygen glucose deprivation/re-oxygenation-induced neuronal cell death by inhibiting NF-κB signaling. Biochem Biophys Res Commun 503:2524–2530. https://doi.org/10.1016/j.bbrc.2018.07.010
Wan P, Su W, Zhang Y et al (2020) LncRNA H19 initiates microglial pyroptosis and neuronal death in retinal ischemia/reperfusion injury. Cell Death Differ 27:176–191. https://doi.org/10.1038/s41418-019-0351-4
Xu L, Zhang Z, Xie T et al (2016) Inhibition of BDNF-AS Provides Neuroprotection for Retinal Ganglion Cells against Ischemic Injury. PloS One 11:e0164941. https://doi.org/10.1371/journal.pone.0164941
Wang L-Q, Zhou H-J (2018) LncRNA MALAT1 promotes high glucose-induced inflammatory response of microglial cells via provoking MyD88/IRAK1/TRAF6 signaling. Sci Rep 8:8346. https://doi.org/10.1038/s41598-018-26421-5
Zhang B, Wang D, Ji T-F et al (2017) Overexpression of lncRNA ANRIL up-regulates VEGF expression and promotes angiogenesis of diabetes mellitus combined with cerebral infarction by activating NF-κB signaling pathway in a rat model. Oncotarget 8:17347–17359. https://doi.org/10.18632/oncotarget.14468
Wang C-F, Zhao C-C, Weng W-J et al (2017) Alteration in long non-coding RNA expression after traumatic brain injury in rats. J Neurotrauma 34:2100–2108. https://doi.org/10.1089/neu.2016.4642
Yu Y, Cao F, Ran Q, Wang F (2017) Long non-coding RNA Gm4419 promotes trauma-induced astrocyte apoptosis by targeting tumor necrosis factor α. Biochem Biophys Res Commun 491:478–485 https://doi.org/10.1016/j.bbrc.2017.07.021
Patel NA, Moss LD, Lee J-Y et al (2018) Long noncoding RNA MALAT1 in exosomes drives regenerative function and modulates inflammation-linked networks following traumatic brain injury. J Neuroinflammation 15:204. https://doi.org/10.1186/s12974-018-1240-3
Yi M, Dai X, Li Q et al (2019) Downregulated lncRNA CRNDE contributes to the enhancement of nerve repair after traumatic brain injury in rats. Cell Cycle Georget Tex 18:2332–2343. https://doi.org/10.1080/15384101.2019.1647024
Cheng S, Zhang Y, Chen S, Zhou Y (2020) LncRNA HOTAIR Participates in Microglia Activation and Inflammatory Factor Release by Regulating the Ubiquitination of MYD88 in Traumatic Brain Injury. J Mol Neurosci MN. https://doi.org/10.1007/s12031-020-01623-7
Jia J, Zhang M, Li Q et al (2018) Long noncoding ribonucleic acid NKILA induces the endoplasmic reticulum stress/autophagy pathway and inhibits the nuclear factor-k-gene binding pathway in rats after intracerebral hemorrhage. J Cell Physiol 233:8839–8849. https://doi.org/10.1002/jcp.26798
Gao M, Fu J, Wang Y (2020) The lncRNA FAL1 protects against hypoxia-reoxygenation- induced brain endothelial damages through regulating PAK1. J Bioenerg Biomembr 52:17–25. https://doi.org/10.1007/s10863-019-09819-2
Zhang X, Zhu X-L, Ji B-Y et al (2019) LncRNA-1810034E14Rik reduces microglia activation in experimental ischemic stroke. J Neuroinflammation 16:75. https://doi.org/10.1186/s12974-019-1464-x
Zheng B, Liu H, Wang R et al (2015) Expression signatures of long non-coding RNAs in early brain injury following experimental subarachnoid hemorrhage. Mol Med Rep 12:967–973. https://doi.org/10.3892/mmr.2015.3474
Shao H-F, Li Z-Z, Zheng X-F et al (2019) Research on the correlation of changes in plasma lncRNA MEG3 with change in inflammatory factors and prognosis in patients with traumatic brain injury. Eur Rev Med Pharmacol Sci 23:4341–4347. https://doi.org/10.26355/eurrev_201905_17940
Ji R-R, Xu Z-Z, Gao Y-J (2014) Emerging targets in neuroinflammation-driven chronic pain. Nat Rev Drug Discov 13:533–548. https://doi.org/10.1038/nrd4334
Zhou Z, Han B, Jin H, et al (2020) Changes in long non-coding RNA transcriptomic profiles after ischemia-reperfusion injury in rat spinal cord. PeerJ 8:e8293. https://doi.org/10.7717/peerj.8293
Jia H, Ma H, Li Z et al (2019) Downregulation of LncRNA TUG1 inhibited TLR4 signaling pathway-mediated inflammatory damage after spinal cord ischemia reperfusion in rats via suppressing TRIL expression. J Neuropathol Exp Neurol 78:268–282. https://doi.org/10.1093/jnen/nly126
Zhou H-J, Wang L-Q, Wang D-B et al (2018) Long noncoding RNA MALAT1 contributes to inflammatory response of microglia following spinal cord injury via the modulation of a miR-199b/IKKβ/NF-κB signaling pathway. Am J Physiol Cell Physiol 315:C52–C61. https://doi.org/10.1152/ajpcell.00278.2017
Zhou J, Li Z, Wu T et al (2020) LncGBP9/miR-34a axis drives macrophages toward a phenotype conducive for spinal cord injury repair via STAT1/STAT6 and SOCS3. J Neuroinflammation 17:134. https://doi.org/10.1186/s12974-020-01805-5
Shao M, Jin M, Xu S et al (2020) Exosomes from long noncoding RNA-Gm37494-ADSCs repair spinal cord injury via shifting microglial M1/M2 polarization. Inflammation 43:1536–1547. https://doi.org/10.1007/s10753-020-01230-z
Li H, Xu Y, Wang G et al (2019) Long non-coding RNA Mirt2 relieves lipopolysaccharide-induced injury in PC12 cells by suppressing miR-429. J Physiol Biochem 75:403–413. https://doi.org/10.1007/s13105-019-00691-7
Zhu S, Zhou Z, Li Z et al (2019) Suppression of LINC00707 alleviates lipopolysaccharide-induced inflammation and apoptosis in PC-12 cells by regulated miR-30a-5p/Neurod 1. Biosci Biotechnol Biochem 83:2049–2056. https://doi.org/10.1080/09168451.2019.1637245
Jiang B-C, Sun W-X, He L-N et al (2015) Identification of IncRNA expression profile in the spinal cord of mice following spinal nerve ligation-induced neuropathic pain. Mol Pain 11:s12990-015–0047. https://doi.org/10.1186/s12990-015-0047-9
Hu J-Z, Rong Z-J, Li M et al (2019) Silencing of lncRNA PKIA-AS1 attenuates spinal nerve ligation-induced neuropathic pain through epigenetic downregulation of CDK6 expression. Front Cell Neurosci 13:50. https://doi.org/10.3389/fncel.2019.00050
Pan X, Shen C, Huang Y et al (2020) Loss of SNHG4 attenuated spinal nerve ligation-triggered neuropathic pain through sponging miR-423-5p. Mediators Inflamm 2020:2094948. https://doi.org/10.1155/2020/2094948
Wu J, Wang C, Ding H (2020) LncRNA MALAT1 promotes neuropathic pain progression through the miR-154-5p/AQP9 axis in CCI rat models. Mol Med Rep 21:291–303. https://doi.org/10.3892/mmr.2019.10829
Pang H, Ren Y, Li H et al (2020) LncRNAs linc00311 and AK141205 are identified as new regulators in STAT3-mediated neuropathic pain in bCCI rats. Eur J Pharmacol 868:172880. https://doi.org/10.1016/j.ejphar.2019.172880
Yan X-T, Lu J-M, Wang Y et al (2018) XIST accelerates neuropathic pain progression through regulation of miR-150 and ZEB1 in CCI rat models. J Cell Physiol 233:6098–6106. https://doi.org/10.1002/jcp.26453
Wei M, Li L, Zhang Y et al (2018) LncRNA X inactive specific transcript contributes to neuropathic pain development by sponging miR-154-5p via inducing toll-like receptor 5 in CCI rat models. J Cell Biochem. https://doi.org/10.1002/jcb.27088
Zhao Y, Li S, Xia N et al (2018) Effects of XIST/miR-137 axis on neuropathic pain by targeting TNFAIP1 in a rat model. J Cell Physiol 233:4307–4316. https://doi.org/10.1002/jcp.26254
Jin H, Du X-J, Zhao Y, Xia D-L (2018) XIST/miR-544 axis induces neuropathic pain by activating STAT3 in a rat model. J Cell Physiol 233:5847–5855. https://doi.org/10.1002/jcp.26376
Xia L-X, Ke C, Lu J-M (2018) NEAT1 contributes to neuropathic pain development through targeting miR-381/HMGB1 axis in CCI rat models. J Cell Physiol 233:7103–7111. https://doi.org/10.1002/jcp.26526
Shen F, Zheng H, Zhou L et al (2019) LINC00657 expedites neuropathic pain development by modulating miR-136/ZEB1 axis in a rat model. J Cell Biochem 120:1000–1010. https://doi.org/10.1002/jcb.27466
Zhang D, Mou J-Y, Wang F et al (2019) CRNDE enhances neuropathic pain via modulating miR-136/IL6R axis in CCI rat models. J Cell Physiol 234:22234–22241. https://doi.org/10.1002/jcp.28790
Chen Z-L, Liu J-Y, Wang F, Jing X (2019) Suppression of MALAT1 ameliorates chronic constriction injury-induced neuropathic pain in rats via modulating miR-206 and ZEB2. J Cell Physiol. https://doi.org/10.1002/jcp.28213
Xu M, Yan Y, Zhu M et al (2020) Effects of long non-coding RNA Gm14461 on pain transmission in trigeminal neuralgia. J Inflamm Lond Engl 17:1. https://doi.org/10.1186/s12950-019-0231-1
Sun W, Ma M, Yu H, Yu H (2018) Inhibition of lncRNA X inactivate-specific transcript ameliorates inflammatory pain by suppressing satellite glial cell activation and inflammation by acting as a sponge of miR-146a to inhibit Nav 1.7. J Cell Biochem 119:9888–9898. https://doi.org/10.1002/jcb.27310
Liu Y, Feng L, Ren S et al (2020) Inhibition of lncRNA DILC attenuates neuropathic pain via the SOCS3/JAK2/STAT3 pathway. Biosci Rep 40. https://doi.org/10.1042/BSR20194486
Xia X, Niu H, Ma Y et al (2020) LncRNA CCAT1 protects astrocytes against OGD/R-induced damage by targeting the miR-218/NFAT5-signaling axis. Cell Mol Neurobiol. https://doi.org/10.1007/s10571-020-00824-3
Yu Y, Zhu M, Zhao Y et al (2018) Overexpression of TUSC7 inhibits the inflammation caused by microglia activation via regulating miR-449a/PPAR-γ. Biochem Biophys Res Commun 503:1020–1026. https://doi.org/10.1016/j.bbrc.2018.06.111
Li J-W, Kuang Y, Chen L, Wang J-F (2018) LncRNA ZNF667-AS1 inhibits inflammatory response and promotes recovery of spinal cord injury via suppressing JAK-STAT pathway. Eur Rev Med Pharmacol Sci 22:7614–7620. https://doi.org/10.26355/eurrev_201811_16375
Peng C, Zhang C, Su Z, Lin D (2019) DGCR5 attenuates neuropathic pain through sponging miR-330-3p and regulating PDCD4 in CCI rat models. J Cell Physiol 234:7292–7300. https://doi.org/10.1002/jcp.27487
Zheng H, Hu S, Cao J et al (2019) Long non-coding RNA TUG1 alleviates LPS-induced injury of PC-12 cells by down-regulating microRNA-127. Exp Mol Pathol 110:104287. https://doi.org/10.1016/j.yexmp.2019.104287
Mariani MM, Kielian T (2009) Microglia in infectious diseases of the central nervous system. J Neuroimmune Pharmacol 4:448–461. https://doi.org/10.1007/s11481-009-9170-6
Yang R, Huang F, Fu J et al (2016) Differential transcription profiles of long non-coding RNAs in primary human brain microvascular endothelial cells in response to meningitic Escherichia coli. Sci Rep 6:38903. https://doi.org/10.1038/srep38903
Sun W, Pei L, Liang Z (2017) mRNA and long non-coding RNA expression profiles in rats reveal inflammatory features in sepsis-associated encephalopathy. Neurochem Res 42:3199–3219. https://doi.org/10.1007/s11064-017-2357-y
Chen Y, Zhang Y, Ye G et al (2020) Knockdown of lncRNA PCAI protects against cognitive decline induced by hippocampal neuroinflammation via regulating SUZ12. Life Sci 253:117626. https://doi.org/10.1016/j.lfs.2020.117626
Wang P (2019) The opening of pandora’s box: an emerging role of long noncoding RNA in viral infections. Front Immunol 9:3138. https://doi.org/10.3389/fimmu.2018.03138
Meng X-Y, Luo Y, Anwar MN et al (2017) Long non-coding RNAs: Emerging and versatile regulators in host-virus interactions. Front Immunol 8:1663. https://doi.org/10.3389/fimmu.2017.01663
Heaton RK, Clifford DB, Franklin DR et al (2010) HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology 75:2087–2096. https://doi.org/10.1212/WNL.0b013e318200d727
Zhou F, Liu X, Zuo D et al (2018) HIV-1 Nef-induced lncRNA AK006025 regulates CXCL9/10/11 cluster gene expression in astrocytes through interaction with CBP/P300. J Neuroinflammation 15:303. https://doi.org/10.1186/s12974-018-1343-x
Ghosh D, Basu A (2009) Japanese encephalitis-a pathological and clinical perspective. PLoS Negl Trop Dis 3:e437. https://doi.org/10.1371/journal.pntd.0000437
Ghoshal A, Das S, Ghosh S et al (2007) Proinflammatory mediators released by activated microglia induces neuronal death in Japanese encephalitis. Glia 55:483–496. https://doi.org/10.1002/glia.20474
Li Y, Zhang H, Zhu B et al (2017) microarray analysis identifies the potential role of long non-coding RNA in regulating neuroinflammation during Japanese encephalitis virus infection. Front Immunol 8:1237. https://doi.org/10.3389/fimmu.2017.01237
Zhao P, Liu S, Zhong Z et al (2018) Analysis of expression profiles of long noncoding RNAs and mRNAs in brains of mice infected by rabies virus by RNA sequencing. Sci Rep 8:11858. https://doi.org/10.1038/s41598-018-30359-z
Ji S, Zhu M, Zhang J et al (2019) Microarray analysis of lncRNA expression in rabies virus infected human neuroblastoma cells. Infect Genet Evol 67:88–100. https://doi.org/10.1016/j.meegid.2018.10.027
Shih R-H, Wang C-Y, Yang C-M (2015) NF-kappaB Signaling Pathways in Neurological Inflammation: A Mini Review. Front Mol Neurosci 8:77. https://doi.org/10.3389/fnmol.2015.00077
Nabavi SM, Ahmed T, Nawaz M et al (2019) Targeting STATs in neuroinflammation: The road less traveled! Pharmacol Res 141:73–84. https://doi.org/10.1016/j.phrs.2018.12.004
Yan Z, Gibson SA, Buckley JA et al (2018) Role of the JAK/STAT signaling pathway in regulation of innate immunity in neuroinflammatory diseases. Clin Immunol 189:4–13. https://doi.org/10.1016/j.clim.2016.09.014
Danan C, Manickavel S, Hafner M (2016) PAR-CLIP: A method for transcriptome-wide identification of rna binding protein interaction sites. Methods Mol Biol Clifton NJ 1358:153–173. https://doi.org/10.1007/978-1-4939-3067-8_10
Ule J, Hwang H-W, Darnell RB (2018) The future of cross-linking and immunoprecipitation (CLIP). Cold Spring Harb Perspect Biol 10. https://doi.org/10.1101/cshperspect.a032243
Khorkova O, Wahlestedt C (2017) Oligonucleotide therapies for disorders of the nervous system. Nat Biotechnol 35:249–263. https://doi.org/10.1038/nbt.3784
Wurster CD, Ludolph AC (2018) Antisense oligonucleotides in neurological disorders. Ther Adv Neurol Disord 11:1756286418776932. https://doi.org/10.1177/1756286418776932
Acknowledgments
Figure 1 was created with BioRender.com. This work is supported by extramural grants from the Government of India, DBT-BT/PR27371/MED/122/122/2017 to VS and AB; DST Grant No. DST-SERB ECR/2017/001953 and DBT-RLS 102/IFD/SAN/3499/2016-17 to VS; OPERA award from BITS Pilani to VS. BS was supported by a scholarship from BITS Pilani and ICMR SRF. Swati acknowledges DST, New Delhi, for providing fellowship (DST/INSPIRE Fellowship/2019/IF190343).
Author information
Authors and Affiliations
Contributions
VS and AB conceived and designed the manuscript. ST, BS, SM, S, AB, and VS wrote the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no conflict of interest.
Ethics Approval and Consent to Participate
Not applicable
Consent for Publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tripathi, S., Shree, B., Mohapatra, S. et al. The Expanding Regulatory Mechanisms and Cellular Functions of Long Non-coding RNAs (lncRNAs) in Neuroinflammation. Mol Neurobiol 58, 2916–2939 (2021). https://doi.org/10.1007/s12035-020-02268-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-020-02268-8