Abstract
Parkinson's disease (PD) is a major neurodegenerative chronic disease, most likely caused by a complex interplay of genetic and environmental factors. Information on various aspects of PD pathogenesis is rapidly increasing and needs to be efficiently organized, so that the resulting data is available for exploration and analysis. Here we introduce a computationally tractable, comprehensive molecular interaction map of PD. This map integrates pathways implicated in PD pathogenesis such as synaptic and mitochondrial dysfunction, impaired protein degradation, alpha-synuclein pathobiology and neuroinflammation. We also present bioinformatics tools for the analysis, enrichment and annotation of the map, allowing the research community to open new avenues in PD research. The PD map is accessible at http://minerva.uni.lu/pd_map.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Parkinson’s disease (PD) is a major neurodegenerative disease, characterized clinically by a range of symptoms, in particular, impaired motor behaviour. The pathogenesis of PD is multi-factorial and age-related, implicating various genetic and environmental factors [1]. Gaps in the understanding of the underlying molecular mechanisms hamper the design of effective disease modifying therapies. Investigation of such a complex disease requires a proper knowledge repository that organizes the rapidly growing PD-related knowledge — a disease map.
The concept of a disease map is relatively new and has found only a limited application in the field of neurodegenerative diseases thus far [2, 3]. Such a map represents diagrammatically interactions between molecular components and pathways reported to play a role in disease pathogenesis and progression. It provides navigation and exploration tools that help the user to locate specific areas of interest and visualize known interactions. Associated analytical tools allow investigators to develop a profound understanding of the disease, detect unexpected interactions and ultimately identify new research hypotheses.
In this paper, we present a PD molecular interaction map that captures and visualizes all major molecular pathways involved in PD pathogenesis. Furthermore, it constitutes a resource for computational analyses and a platform for community level collaborations [4, 5] (see Fig. 1). We also present how a set of bioinformatics tools applied to the map can facilitate in-depth knowledge extraction and continuous curation.
The concept of Parkinson's disease map and its possibilities. The PD map is a knowledge repository bringing together different molecular mechanisms and pathways considered to be the key players in the disease. The current focus of the map is illustrated by the pieces in the “PD puzzle” These modules include synaptic and mitochondrial dysfunction, failure of protein degradation systems, α-synuclein pathobiology and misfolding, and neuroinflammation. Processes important in PD-associated neurodegeneration, such calcium homeostasis or apoptosis, are discussed within their appropriate context in the main text, and included into the PD map pathways. The PD map is represented as a graph constructed with all gene-regulatory protein and metabolic interactions extracted from published data. Currently the map has 2,285 elements and 989 reactions supported by 429 articles and 254 entries from publicly available bioinformatic databases. It is compliant with standardized graphical representation, Systems Biology Graphical Notation (SBGN) [265]. This standardized representation of the map could become a common language for the PD research community to discuss disease-related molecular mechanisms [5]. Detailed contents of the PD map are accessible at http://minerva.uni.lu/MapViewer/map?id=pdmap (Online resource 1) as an SBML file (Online resource 2) and in Payao [264]. The map can be updated with information from the PD research community, as well as by searching bioinformatics databases. Exploration and analysis of the content has the potential to broaden knowledge on the molecular processes in PD, generate of new hypotheses on disease pathogenesis, or prioritize the most interesting areas and molecules for investigation. Approaches to facilitate this knowledge acquisition process are discussed in detail in the section “Annotation, enrichment and Analysis of the PD Map”
The paper is divided into two parts. In the first part, we review the pathways implicated in PD, with a focus on synaptic and mitochondrial dysfunction, α-synuclein pathobiology, failure of protein degradation systems, neuroinflammation and apoptosis. In the second part of the paper, we demonstrate how the PD map interfaces with bioinformatics tools and databases for its content annotation, enrichment with experimental results, and analysis of its complex structure and dynamics. The PD map is accessible under http://minerva.uni.lu/pd_map (Online resource 1), as a SBML file (Online resource 2), and Payao, a community platform for pathway model curation [264].
Neurodegeneration in Parkinson’s Disease Arises from Dysregulation of Interlinked Molecular Pathways
The major pathological feature of PD is the progressive degeneration of the nigrostriatal system, leading to the loss of dopaminergic (DA) neurons in the substantia nigra pars compacta (SNpc) [6]. The degeneration of the nigrostriatal pathway and subsequent loss of striatal dopamine contributes to the cardinal clinical motor symptoms: tremor, rigidity, bradykinesia and postural instability [7]. Although treatments such as dopamine substitution and deep brain stimulation alleviate many of the motor symptoms, there is no disease-modifying therapy preventing the progressive loss of DA neurons [8].
Susceptibility for PD is modulated by various environmental factors [9–13], genetic predisposition or risk factors [14] and epigenetic alterations [15, 16].Footnote 1 Exposure to pesticides and industrial agents has been associated with an increased risk for PD [17, 18], but to date none of these agents have been consistently identified as a causal factor for PD [19]. It is known that exposure to inhibitors of mitochondrial respiration [20–25] are sufficient to induce PD symptoms in humans and DA neurodegeneration in animal models.
In this paper, we focus on DA neurons as a major point of convergence in PD disease pathways. However, pathogenic pathways leading to the demise of DA neurons may impact any neuronal population affected in PD, including those of the autonomic ganglia [26, 27]. The demise of these populations may contribute to a range of PD-typical non-motor symptoms hampering the life of PD patients, such as constipation and dysautonomia (ganglia of autonomous nervous system), cognitive decline and REM sleep behaviour (cholinergic neurons of the nucleus basalis of Meynert, noradrenergic coeruleus–subcoeruleus complex), depression and apathy (serotinergic caudal raphe nuclei, cholinergic gigantocellular reticular nucleus) [28, 29].
Vulnerability and Preferential Loss of Midbrain Dopaminergic Neurons
SNpc DA neurons are the most vulnerable population of neurons in PD. It has been suggested that their loss is multifactorial and related to the characteristic features of these cells: complex morphology, high energy demand, high calcium flux, and dopamine metabolism [30]. Consequently, these neurons are particularly susceptible to various stressors, which contribute to their preferential loss (see Fig. 2).
Pathways implicated in PD and their relationship to susceptibility factors of SNpc DA neurons. The black arrows represent direct molecular interactions between the dysregulated pathways. Red arrows denote pathways affected by or generating ROS. Dashed lines represent indirect associations of these pathways and neurodegeneration. Susceptibility factors of SNpc DA neurons associated with a given pathway are indicated by their corresponding symbols
SNpc DA neurons have one of the longest yet most dense arborisation of all neurons [31, 32]. They project to the striatum, providing it with DA [33, 34]. These neurons have long, thin, mostly unmyelineated axons [35] and up to 150,000 presynaptic terminals per neuron [30]. The high energy demand required to support synaptic activity, compensation for the potential risk of depolarization in the unmyelinated membrane, and axonal transport over long distances put a huge burden on the mitochondria. Interestingly, toxins that perturb the energy production and the axonal transport of mitochondria [36], cause parkinsonism in humans and preferential loss of DA neurons in animal models [22, 36, 37]. Finally, the large number of synapses increases the risk for local α-synuclein (α-syn) misfolding (see sections “Synaptic Dysfunction” and “α-Synuclein Misfolding and Pathobiology”).
SNpc DA neurons can fire autonomously and have specific calcium L-type Cav 1.3 channels that regulate this pacemaking activity [38, 39]. The resulting high intracytosolic Ca2+ concentrations induce cellular stress, elevate the levels of reactive oxygen species (ROS), and increase demand for calcium buffering, which is handled by the endoplasmic reticulum (ER) and the mitochondria. Maintaining proper calcium homeostasis in such an environment increases again the energy needs. In contrast, neighbouring dopamine neurons in the ventral tegmental area use Na+ channels for pacemaking and are relatively spared in PD [37].
Cytosolic DA also contributes to the vulnerability of DA neurons, primarily because its metabolism induces oxidative and nitrative stress in an age-dependent manner [40–42]. Neurotoxicity of DA increases with its concentration, which is thought to be regulated by Ca2+ concentration [43]. Additionally, dopamine metabolism is involved in a number of PD-associated pathways, as it can impair synapse function, inhibit protein degradation and disturb mitochondrial dynamics by inhibiting the function of Parkin.
Ageing, the primary risk factor for PD, especially affects DA neurons (see Fig. 2). α-Syn accumulation increases with age in the SNpc and correlates with the loss of DA neurons in non-human primates [42]. This could be linked to the age-related impairment of the two protein degradation systems: the ubiquitin–proteasome system (UPS) [42] and the autophagy–lysosome system [44]. ROS accumulate in an ageing brain [42, 45], partially due to mitochondria dysfunction, as mitophagyFootnote 2 is decreased with ageing [45, 46]. Finally, the threshold required to trigger a neuroinflammatory response may decrease with age, since glial activation in SNpc increases in the ageing brain [42, 47].
Synaptic Dysfunction
The main function of a synapse is to establish a connection between neurons allowing communication via chemical or electric signals. The synapse has emerged as a neuronal structure highly susceptible to a variety of chronic insults [48–51]. Below, we discuss the increasing evidence indicating that synapses are also affected in PD, and that their dysfunction and demise contributes to the disease.
α-Syn is a presynaptic protein. Point mutations, duplications or triplications of its gene are associated with familial PD [52–54]. In cultured neurons, it transiently associates with synaptic vesicles prior to neurotransmitter release, upon which it rapidly redistributes to the cytosol [55]. Association of α-syn with the synaptic vesicle may occur through its binding to SNARE complex proteins [56], and, as shown in mice, α-syn positively influences functional SNARE levels [57]. Similarly, upregulation of α-syn in synapses and cell somas of cultured neurons protects against oxidative stress [58]. However, the protective effect of α-syn is limited to a narrow concentration range, since high levels of α-syn cause familial PD [53]. Even modest overexpression of α-syn has been reported to markedly inhibit neurotransmitter release [59]. Also, α-syn forms potentially pathogenic micro-aggregates in the synapse [60]. Another protein involved familial and sporadic PD, LRRK2, is also present in the synapse. Its experimentally induced upregulation or knockdown impairs the dynamics of synaptic vesicle release and recycling [61, 62]. However, the influence of mutated or dysfunctional LRRK2 on these processes in PD remains to be investigated.
A number of other PD-related pathological events might affect synapses. Synapses of the nigrostriatal pathway, with their high level of α-syn and dopamine, are likely to be the major site of the formation of toxic adducts of α-syn and oxidized DA [40, 63, 64]. Furthermore, the energy demands of synapses may be compromised by dysfunctional mitochondrial respiration, turnover, or axonal transport [65]. Locally dysfunctional protein degradation and turnover may directly affect synaptic function and plasticity [66].
Mitochondrial Dysfunction
Mitochondria are highly dynamic organelles essential for a range of cellular processes including ATP production, ROS management, calcium homeostasis, and control of apoptosis. The maintenance of mitochondrial homoeostasis by mitophagy involves multiple factors ranging from the control of mitochondrial fusion and fission to mitochondrial motility [67]. These processes are strongly related to proteins involved in familial and sporadic PD [65, 68, 69].
A number of proteins associated with familial PD are related to mitochondrial function [70], with PINK1 and Parkin playing a particularly important role. Control of mitochondrial turnover and protection against oxidative stress are mediated via the kinase activity of PINK1 targeting Parkin [71], HTRA2 [72] and TRAP1[73] proteins. In turn, mitophagy is driven by PINK1-mediated translocation of Parkin from the cytosol to mitochondria [71, 74]. Importantly, both mitophagy [75, 76] and transcriptional control of mitochondrial biogenesis [77–79] depend on the E3 ubiquitin ligase activity of Parkin.
Familial PD genes are also implicated in ROS production by mitochondria. Mitochondrial respiration and calcium balance are perturbed by PINK1 deficiency [80, 81]. The resulting reduced mitochondrial calcium capacity and increased ROS could lower the threshold for mitochondrial outer membrane permeabilization (MOMP) and thereby increase the vulnerability for cell death [80]. Additional detrimental downstream effects of excessive ROS are mitochondrial DNA damage and inflammation [65, 82]. It has been suggested that DJ1 works in parallel to the PINK1–Parkin pathway to maintain mitochondrial function in the presence of an oxidative environment [83]. DJ1 was shown to interact with a mitochondrial protein mortalin, which maintains mitochondrial homeostasis and antagonizes oxidative stress injury [84]. Remarkably, Parkin overexpression has been demonstrated to prevent mitochondrial dysfunction caused by a mortalin knockdown [85].
Mitochondrial trafficking is necessary for proper energy supply. This process is particularly demanding in long axons of DA neurons. Recent findings suggest that mitochondrial transport may be affected in PD. Axonal transport of mitochondria along the microtubules is directly influenced by PINK1 through its supporting role in the kinesin motor complex [86]. Also, PINK1 and Parkin may play an important role in the process of quarantining the damaged mitochondria prior to their clearance [87]. However, the role of PINK1 in the dynamics of mitochondrial trafficking is not yet fully understood [88]. Mitochondrial trafficking may also be impaired by Parkin, α-syn, or LRRK2 as they modulate microtubule stability [89–92], or by formation of α-syn aggregates [93].
Finally, other proteins associated with familial PD have recently been linked to mitochondrial pathways. UCHL1-mediated cell death can be attenuated by mitochondrial protein HTRA2 [94], ATP13A2 regulates mitochondrial bioenergetics through macroautophagy [95], VPS35 mediates vesicle transport between mitochondria and peroxisomes [96], and EIF4G1 is involved in stress related protection of mitochondria [97].
Failure of Protein Degradation Systems
In long-lived post-mitotic cells, such as neurons, the degradation systems assuring the removal of damaged, dysfunctional cellular structures play a key role in cellular homeostasis. These degradation systems are involved in the clearance of defective cellular structures such as misfolded or damaged proteins, and dysfunctional organelles such as defective mitochondria [98]. The two major degradation systems are the UPS and the autophagy–lysosome system. The complex machinery and biology of these two systems have been extensively reviewed elsewhere [66, 99–101]. The dysfunction of clearance systems, especially in the synapse, can lead to the accumulation of α-syn and defective mitochondria. These, in turn, can interfere with proper synaptic function, lead to the formation of toxic assemblies or aggregates, or impair energy metabolism and cause oxidative stress.
Genetic and pathological evidence strongly indicate the involvement of defective clearance systems in PD [102–104]. Interestingly, patients with Gaucher’s disease, a lysosomal storage disorder [105, 106], have an increased risk for PD and accumulate α-syn in their brains [107]. Mutated forms of α-syn have been reported to inhibit their own degradation by chaperone-mediated autophagy (CMA), while DA-modified α-syn also blocks CMA degradation of other proteins [103]. Finally, pathological observations in PD autopsy brains and brains of PD animal models show an increased number of autophagy vacuoles and other autophagy markers [108, 109]. Interestingly, neurons containing Lewy bodies (LB) were shown to have decreased UPS and lysosomal markers [110].
While this evidence demonstrates the involvement of cellular clearance mechanisms in PD, it is unclear whether that involvement is primarily beneficial or detrimental. It has been argued that exaggerated clearance activity may contribute to neuronal injury [111, 112]. The predominant view, however, is that the removal of abnormal proteins and organelles is neuroprotective [102, 113–118].
α-Synuclein Misfolding and Pathobiology
The pathobiology of α-syn is implicated in a number of pathways involved in PD. α-Syn is an intrinsically disordered protein [119], which can spontaneously and dynamically adopt either physiological or misfolded conformations. The latter contains β-sheet structure, which promotes oligomerisation and fibrilisation [120–122]. High-order oligomeric and pre-fibrillar forms are thought to be cytotoxic, while fibrillar and aggregated forms may be harmless, detoxified depositions [119, 123]. This is still controversial, since familial PD α-syn mutants promote both misfolding and aggregation of α-syn, suggesting a pathological role of this process [103, 121, 124, 125].
Mutated, misfolded or overexpressed α-syn is involved in a number of pathways associated with degeneration of SNpc DA neurons. It is thought to impair synapse function [126–129] and to affect the respiration, morphology and turnover of mitochondria [130–134]. Axonal transport might be impaired by misfolded α-syn through perturbation of microtubule assembly [135–137], especially together with MAPT protein [138–143]. Also, oligomers of mutant α-syn induce chronic ER stress [125, 144], which seems to precede actual neurodegeneration [145]. Finally, α-syn degradation by CMA [146] might be perturbed by mutated or dopamine-modified α-syn [103, 146, 147]. Reduction of lysosomal activity by α-syn overexpression might lead to α-syn accumulation [148], suggesting a vicious loop of CMA deficiency and α-syn misfolding. The proteasome system has also been reported to be inhibited either by α-syn mutants [149–151], or oligomers [152].
Recent studies suggest that α-syn aggregates spread between cells and that this contributes to the PD disease process [123, 153]. This hypothesis is supported by reports of protein inclusions detected in previously unaffected DA neurons grafted into the striatum of PD patients [154–156]. The existence of a neuron-to-neuron transfer mechanism for misfolded α-syn has been shown in cell culture, primary mouse neurons and mouse models [157–159]. Moreover, it was observed that different types of cellular stress associated with PD pathogenesis, such as misfolded protein accumulation [160], proteasomal and mitochondrial dysfunction [161], are able to increase secretion of α-syn and its aggregates.
It has been shown that exogenous α-syn preformed fibrils might promote the aggregation of endogenous α-syn in neuronal cells [158, 159, 162] impairing neural function [158, 159]. Taken together, these results suggest that misfolded α-syn can be secreted and taken up, introducing additional cellular stress and promoting further protein misfolding.
Neuroinflammation
Neuroinflammation and chronic activation of the immune system are pathological processes associated with all chronic neurodegenerative diseases, such as PD, AD or multiple sclerosis [163]. Although the involvement of the adaptive immune system in PD-related neuroinflammation has been suggested [164, 165], in particular in the context of α-syn and neuromelanin [166, 167], current research of neuroinflammation in PD focuses primarily on the innate immune system. Of particular interest are microgliaFootnote 3 [168] and astrocytesFootnote 4 [169, 170].
Microglia constantly explore and monitor the local environment [171, 172], modulating the response of the immune system in relation to the level of their perturbation. At the first sign of stress, they produce and release anti-inflammatory cytokines and supportive growth factors [168]. Neurons play an active role in regulating the microglial response. Many of their products inhibit microglia activation by binding to specific microglial receptors [173–177].
The SNpc is a brain region that may be especially vulnerable to elevated neuroinflammation. The SNpc contains more microglia [178] and less astrocytes than other brain regions [179]. With a high microglial density promoting the inflammatory response, and low astroglial density to downregulate it, neuroinflammation in the SNpc may be particularly strong. Moreover, SNpc neurons contain neuromelanin, which has been shown to activate microglia [180] and could be another factor promoting neuroinflammation.
The response of glial cells in the context of PD has been studied in humans, animal models and cell cultures. The presence of reactive microglia in human post-mortem brain tissue has been reported in PD patients [181] and in people exposed to MPTP [182]. In animal models of PD, microglial activation has been studied in primates [183], mice [184] and rats [185], supporting the notion that neuroinflammation is intrinsically associated with the PD pathological process. In cellular co-cultures of neuronal cells and microglia, neuronal injury drives microglia activation, which in turn enhances neurodegeneration [186].
In vitro systems demonstrate that the delicate balance between protective and detrimental effects of glial response might be disrupted by PD-related stress factors. Microglia can detect misfolded α-syn [187, 188] and increase neurotoxicity by producing ROS and pro-inflammatory cytokines [189, 190]. In turn, activated microglia expressing LRRK2 with a PD-related mutation produce more pro-inflammatory cytokines than corresponding cells expressing WT LRRK2 [191]. Deficiency in Parkin may indirectly promote microglia activation by increasing neuronal vulnerability for inflammation-related stress [192] and disturbing the neuron–microglia balance. Finally, DJ-1 deficiency in astrocytes might contribute to neurodegeneration by deregulating their neuroinflammatory response [193].
In summary, many in vitro PD models indicate a detrimental role of microglia. However, the situation in vivo is less clear, even though protective effects of anti-inflammatory compounds such as minocycline have been reported in models of PD [194].
Neuronal Death Through Apoptosis-Related Mechanisms
Degeneration of DA neurons is the final consequence of dysregulated cellular processes, leading to neuronal death [195]. Neurodegeneration by apoptosis typically proceeds through one of two signalling cascades, termed the intrinsic and extrinsic pathways [196, 197].
The intrinsic pathway can be induced by intracellular stress, leading to MOMP that is controlled through proteins of the BCL-2 family. As a result, cytochrome c is released from the mitochondrial intermembrane space, leading to formation of an apoptosome and subsequent execution of apoptosis by activation of caspases 3 and 7. Studies in animal models of PD suggest the BCL-2 family is a key target for attenuating neurodegeneration of DA neurons [198–202].
Additionally, an important link between PD and the activation of apoptosis comes from studies investigating the roles of familial PD genes. It has been shown that disease-related LRRK2 mutations R1441C, Y1699C and G2019S promote mitochondria-dependent apoptosis [203]. PINK1 and Parkin, in turn, protect against stress-induced cytochrome c release, while their mutations might fail to attenuate basal neuronal pro-apoptotic activity [204–206]. Importantly, failure of the protein degradation system could also contribute to apoptosis via the intrinsic pathway. It has been proposed that lysosome membrane permeabilization is induced by ROS and occurs upstream and downstream of MOMP [207, 208]. Finally, DA-mediated activation of the intrinsic pathway may contribute to selective DA neuron degeneration [209].
The extrinsic apoptosis pathway is activated by extracellular signalling, and diverges into two sub-pathways: one directly activating caspase 3 and 7, the other causing MOMP. Neuroinflammation could be a major factor in this process [165, 187, 210], promoting neuronal apoptosis either by oxidative insults [211] or by pro-inflammatory cytokines [212, 213].
In summary, both apoptosis pathways appear to be the convergence point of different pathways dysregulated in PD. Still, therapeutic interventions may be most efficacious in maintaining DA neuron functionality if aimed at the upstream events of apoptosis. Indeed, as the apoptotic process is advanced, the intervention may be too late.
Annotation, Enrichment and Analysis of the PD Map
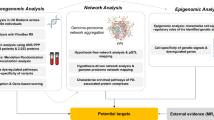
Dysregulated pathways implicated in PD are strongly coupled, and their interconnections need to be represented in an integrated and comprehensive way to be studied efficiently. Our PD map allows navigation through information on PD-associated mechanisms, and constitutes an interface to well-established tools and methods for updating, enriching, and analysing its contents (see Fig. 3).
The workflow and an illustration of PD map functionalities. a The PD map can be automatically enriched with experimental data and annotated with information from text and bioinformatics databases. The analysis requires no external data sources. b A simplified representation of the PD map is given, with circles (nodes) as map elements and lines (edges) as interactions (uni- or bi-directional). Enrichment: green and red nodes represent up- and downregulated genes, respectively, derived from experimental data; a predicted new map component (square) shares interaction with existing map components (dashed lines) and matches their expression profile. Analysis: nodes with high centrality (blue) play a key role in the network topology and indicate molecules regulating many processes; detection of paths (thick lines) highlights non-trivial relations between elements of a biological process; kinetic modelling reveals temporal dependencies between behaviour of different molecules. Annotation: text mining of PD-related articles suggests new interactions in the map (thick dashed line) and facilitates handling of a huge number of publications; each map element is annotated with information from various bioinformatics databases giving easy access to information about interesting elements
Annotation of the PD Map Using Bioinformatic Databases
We have enriched the elements of the PD map using a number of publicly available databases [214–230]. Information on official gene symbol, synonyms, description and chromosomal location; association with biological processes and diseases; or molecular interacting partners have been embedded within the map. Annotation of the contents of the PD map facilitates the knowledge exploration by providing additional information about map elements and their interactions, and is easily accessible online (see Fig. 3b for illustration and Online resource 1 for details).
Exploration of the Map Using Integrative Expression Analysis
Recently, a variety of PD-related large-scale datasets have become publicly available, including microarray gene expression data for human post mortem samples from different regions of the brain [231–234], human whole-blood samples [235], and samples from animal [236] and cell culture models [237]. This experimental data can be visualized on the PD map or used to predict new map elements and interactions [238–240].
Visualization of gene deregulation in PD-related microarray datasets [231–234] is possible via a variety of methods for candidate disease gene or protein prioritizations [241–244]. We have chosen an approach combining significance scores [245] for differentially expressed genes in multiple studies, and prepared a colour-coded version of the map highlighting upregulation in green and downregulation in red (see the online PD map and Online resource 1). This gives an immediate overview of pathways that are affected by dysregulated genes.
One of the major advantages of the PD map is the possibility to predict new elements and interactions on the basis of the map contents and experimental data. To achieve this, publicly available human molecular interaction data [242] are obtained for the PD map elements, extending the number of interactions. Then, an automated, graph-theoretic approach [243] prioritizes candidate disease proteins that are densely interconnected in the extended PD map, and whose gene expression levels are differentially expressed in the microarray PD samples. We have combined the abovementioned experimental microarray [231–234] and protein-protein interaction data [242] to demonstrate the usage of this approach. The extended PD map containing the prioritized new proteins can be found in Online resource 1.
PD Map as a Network: from Structural Analysis to Kinetic Models
The PD map is a large, complex network integrating metabolic reactions, gene regulation, and signalling processes. Exploring how different elements in the network may influence each other is difficult and non-intuitive. Graph-theoretical methods aim to bridge the gap between our understanding of the role of single elements in a cellular network and the properties of this network as whole [246, 247]. These methods aim to identify key network nodes (genes, proteins), edges (molecular interactions) or modulesFootnote 5 (subnetworks) [246].
Basic properties of individual network elements such as node centralityFootnote 6 indicate their global role in the whole network. In turn, analysis of inter-modular communicationf in the network indicates a how given molecule, complex or interaction can affect communication between modules [248, 249]. More advanced, functional dependencies between elements in the network can be revealed by methods exploring the relationships of all possible paths between network elements and selected molecular dysfunctions [250] (see Fig. 3b for illustration). Examples of network analysis applied to the PD map can be found in Online resource 1.
Most of the connections on the PD map depict real physical interactions between biomolecules. Currently, the PD map contains no information on kinetics of these interactions; however, they can be easily assigned and analysed mathematically [251]. The PD map is compliant with the Systems Biology Markup Language (SBML) standard [252], used by commonly available software to build kinetic models and run simulations [253]. Although assigning kinetic parameters to all interactions in the PD map is a truly challenging task, describing kinetics of a certain process representing a module within the PD map is feasible. In-depth analysis of the dynamics of a process can provide insight as to how elements of the process change quantitatively and over time (see Fig. 3b for illustration) and assess their influence on the related components in the map. This can lead to new hypotheses that are impossible to discover by visual examination or analysis of static network topology [254]. There are many successful examples, where similar bottom–up modelling has been applied to neurobiology related systems [255–258].
In summary, structural network analysis allows for detection of elements key to PD pathogenesis represented in the map. This can serve as a basis for new hypotheses and prioritization of targets for further investigation.
Conclusions and Perspectives
PD is a neurodegenerative disease involving a complex interplay of environmental and genetic factors. It becomes increasingly important to develop new approaches to organize and explore the exploding knowledge of this field. The PD map is a computer-based knowledge repository, representing diagrammatically molecular mechanisms of PD in a structured and standardized way. It can be linked to bioinformatics tools facilitating exploration and updating the contents of the map using bioinformatic annotations.
The main insights into molecular pathology of PD come from studies on familial PD and GWAS. In the future, massive use of next-generation sequencing will provide even more data that might contribute to PD. The PD map facilitates integration and visualization of large experimental datasets, allowing analyzing them in the context of disease mechanisms.
Discovering causal factors of PD pathogenesis is difficult because molecular pathways dysregulated in neurodegeneration are interconnected and influence each other. Analysis of the topology and dynamics of molecular interactions within and across different pathways represented in the PD map may help to uncover key factors in PD pathology. For instance, the role of neuroinflammation in the pathological cascade in PD remains unclear, while the apoptosis, clearly a downstream factor of PD, involves other mechanisms implicated in PD, like protein degradation or mitochondrial quality control. Consequential steps of PD pathology can be elucidated by the global, systems level analysis of all implicated factors.
The map has reached substantial size and complexity. Keeping it up-to-date and refining it with limited resources will be a challenge. We foresee the PD map as a crowd-sourcing project, where an interested and knowledgeable research community is engaged in solving a problem [259–262], similar to WikiPathways or Payao [263, 264], but focused on disease-related mechanisms. Thus, the PD community will easily explore and curate the PD-related knowledge in an online manner, ensuring that individual contributions are recognized.
Notes
Epigenetic alterations — secondary, environmentally induced changes of gene expression.
Mitophagy — autophagy of mitochondria.
Microglia — the most abundant of the resident macrophage populations in the CNS.
Astrocytes — glial cells that play a supportive role for neurons and modulate microglia response.
Module — in the PD map by a module (subgraph, subnetwork) we understand elements and interactions participating in the same pathway or serve similar biological function. Inter-modular communication denotes all interactions linking different modules.
Node centrality — a measure describing how important a given node is for the connectivity of the entire network.
References
Obeso JA, Rodriguez-Oroz MC, Goetz CG et al (2010) Missing pieces in the Parkinson’s disease puzzle. Nat Med 16:653–661. doi:10.1038/nm.2165
Caron E, Ghosh S, Matsuoka Y et al (2010) A comprehensive map of the mTOR signaling network. Mol Syst Biol 6:453. doi:10.1038/msb.2010.108
Mizuno S, Iijima R, Ogishima S et al (2012) AlzPathway: a comprehensive map of signaling pathways of Alzheimer’s disease. BMC Syst Biol 6:52. doi:10.1186/1752-0509-6-52
Ghosh S, Matsuoka Y, Asai Y et al (2011) Software for systems biology: from tools to integrated platforms. Nat Rev Genet 12:821–832
Kitano H, Ghosh S, Matsuoka Y (2011) Social engineering for virtual “big science” in systems biology. Nat Chem Biol 7:323–326
Lees AJ, Hardy J, Revesz T (2009) Parkinson’s disease. Lancet 373:2055–2066. doi:10.1016/S0140-6736(09)60492-X
Jankovic J (2008) Parkinson’s disease: clinical features and diagnosis. J Neurol Neurosurg Psychiatry 79:368–376. doi:10.1136/jnnp.2007.131045
Meissner WG, Frasier M, Gasser T et al (2011) Priorities in Parkinson’s disease research. Nat Rev Drug Disc 10:377–393. doi:10.1038/nrd3430
Dick FD, De Palma G, Ahmadi A et al (2007) Environmental risk factors for Parkinson’s disease and parkinsonism: the Geoparkinson study. Occup Environ Med 64:666–672. doi:10.1136/oem.2006.027003
Goldman SM, Tanner CM, Oakes D et al (2006) Head injury and Parkinson’s disease risk in twins. Ann Neurol 60:65–72. doi:10.1002/ana.20882
Ritz B, Ascherio A, Checkoway H et al (2007) Pooled analysis of tobacco use and risk of Parkinson disease. Arch Neurol 64:990–997. doi:10.1001/archneur.64.7.990
Hu G, Bidel S, Jousilahti P et al (2007) Coffee and tea consumption and the risk of Parkinson’s disease. Mov Disord 22:2242–2248. doi:10.1002/mds.21706
Saunders-Pullman R (2003) Estrogens and Parkinson disease: neuroprotective, symptomatic, neither, or both? Endocrine 21:81–87. doi:10.1385/ENDO:21:1:81
Klein C, Westenberger A (2012) Genetics of Parkinson’s disease. Cold Spring Harb Perspect Med 2:a008888. doi:10.1101/cshperspect.a008888
Marques SCF, Oliveira CR, Pereira CMF, Outeiro TF (2011) Epigenetics in neurodegeneration: a new layer of complexity. Prog Neuro-psychopharmacol Biol Psychiatry 35:348–355. doi:10.1016/j.pnpbp.2010.08.008
Migliore L, Coppedè F (2009) Genetics, environmental factors and the emerging role of epigenetics in neurodegenerative diseases. Mutat Res 667:82–97. doi:10.1016/j.mrfmmm.2008.10.011
Schapira AHV (2011) Aetiopathogenesis of Parkinson’s disease. J Neurol 258:S307–S310. doi:10.1007/s00415-011-6016-y
Priyadarshi A, Khuder SA, Schaub EA, Priyadarshi SS (2001) Environmental risk factors and Parkinson’s disease: a metaanalysis. Environ Res 86:122–127. doi:10.1006/enrs.2001.4264
Dick FD (2006) Parkinson’s disease and pesticide exposures. Br Med Bull 79–80:219–231. doi:10.1093/bmb/ldl018
Davis GC, Williams AC, Markey SP et al (1979) Chronic Parkinsonism secondary to intravenous injection of meperidine analogues. Psychiatry Res 1:249–254
Langston JW, Ballard P, Tetrud JW, Irwin I (1983) Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science 219:979–980
Betarbet R, Sherer TB, MacKenzie G et al (2000) Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci 3:1301–1306. doi:10.1038/81834
Thiruchelvam M, Richfield EK, Baggs RB et al (2000) The nigrostriatal dopaminergic system as a preferential target of repeated exposures to combined paraquat and maneb: implications for Parkinson’s disease. J Neurosci 20:9207–9214
Gash DM, Rutland K, Hudson NL et al (2008) Trichloroethylene: Parkinsonism and complex 1 mitochondrial neurotoxicity. Ann Neurol 63:184–192. doi:10.1002/ana.21288
Goldman SM (2010) Trichloroethylene and Parkinson’s disease: dissolving the puzzle. Expert Rev Neurother 10:835–837. doi:10.1586/ern.10.61
Braak H, Del Tredici K, Rüb U et al (2003) Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging 24:197–211
Poulopoulos M, Levy OA, Alcalay RN (2012) The neuropathology of genetic Parkinson’s disease. Mov Disord 000:1–12. doi:10.1002/mds.24962
Wolters EC (2009) Non-motor extranigral signs and symptoms in Parkinson’s disease. Parkinsonism & Relat Disord 15(Suppl 3):S6–S12. doi:10.1016/S1353-8020(09)70770-9
Ferrer I, López-Gonzalez I, Carmona M et al (2012) Neurochemistry and the non-motor aspects of PD. Neurobiol Dis 46:508–526. doi:10.1016/j.nbd.2011.10.019
Sulzer D (2007) Multiple hit hypotheses for dopamine neuron loss in Parkinson’s disease. Trends Neurosci 30:244–250. doi:10.1016/j.tins.2007.03.009
Matsuda W, Furuta T, Nakamura KC et al (2009) Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. J Neurosci 29:444–453
Matsuda W (2012) Imaging of dopaminergic neurons and the implications for Parkinson’s disease. Syst Biol Parkinson’s Dis. doi:10.1007/978-1-4614-3411-5
Parent M, Parent A (2006) Relationship between axonal collateralization and neuronal degeneration in basal ganglia. J Neural Transm Suppl 85–8
Schmitz Y, Luccarelli J, Kim M et al (2009) Glutamate controls growth rate and branching of dopaminergic axons. J Neurosci 29:11973–11981. doi:10.1523/JNEUROSCI.2927-09.2009
Braak H, Bohl JR, Müller CM et al (2006) Stanley Fahn Lecture 2005: the staging procedure for the inclusion body pathology associated with sporadic Parkinson’s disease reconsidered. Mov Disord 21:2042–2051. doi:10.1002/mds.21065
Kim-Han JS, Antenor-Dorsey JA, O’Malley KL (2011) The Parkinsonian mimetic, MPP+, specifically impairs mitochondrial transport in dopamine axons. J Neurosci 31:7212–7221. doi:10.1523/JNEUROSCI.0711-11.2011
Dauer W, Przedborski S (2003) Parkinson’s disease: mechanisms and models. Neuron 39:889–909
Chan CS, Guzman JN, Ilijic E et al (2007) “Rejuvenation” protects neurons in mouse models of Parkinson’s disease. Nature 447:1081–1086. doi:10.1038/nature05865
Surmeier DJ, Guzman JN, Sanchez-Padilla J, Schumacker PT (2011) The role of calcium and mitochondrial oxidant stress in the loss of substantia nigra pars compacta dopaminergic neurons in Parkinson’s disease. Neuroscience 198:221–231
Asanuma M, Miyazaki I, Ogawa N (2003) Dopamine- or l-DOPA-induced neurotoxicity: the role of dopamine quinone formation and tyrosinase in a model of Parkinson’s disease. Neurotox Res 5:165–176
Cantuti-Castelvetri I, Shukitt-Hale B, Joseph JA (2003) Dopamine neurotoxicity: age-dependent behavioral and histological effects. Neurobiol aging 24:697–706
Collier TJ, Kanaan NM, Kordower JH (2011) Ageing as a primary risk factor for Parkinson’s disease: evidence from studies of non-human primates. Nat Rev Neurosci 12:359–366. doi:10.1038/nrn3039
Mosharov EV, Larsen KE, Kanter E et al (2009) Interplay between cytosolic dopamine, calcium, and alpha-synuclein causes selective death of substantia nigra neurons. Neuron 62:218–229. doi:10.1016/j.neuron.2009.01.033
Rubinsztein DC, Mariño G, Kroemer G (2011) Autophagy and aging. Cell 146:682–695. doi:10.1016/j.cell.2011.07.030
Mammucari C, Rizzuto R (2010) Signaling pathways in mitochondrial dysfunction and aging. Mech Ageing Dev 131:536–543. doi:10.1016/j.mad.2010.07.003
Green DR, Galluzzi L, Kroemer G (2011) Mitochondria and the autophagy–inflammation–cell death axis in organismal aging. Science 333:1109–1112. doi:10.1126/science.1201940
Venkateshappa C, Harish G, Mythri RB et al (2012) Increased oxidative damage and decreased antioxidant function in aging human substantia nigra compared to striatum: implications for Parkinson’s disease. Neurochem Res 37:358–369. doi:10.1007/s11064-011-0619-7
Mallucci GR (2009) Prion neurodegeneration: starts and stops at the synapse. Prion 3:195–201
Sisková Z, Sanyal NK, Orban A et al (2010) Reactive hypertrophy of synaptic varicosities within the hippocampus of prion-infected mice. Biochem Soc Trans 38:471–475
Masliah E (1998) Mechanisms of synaptic pathology in Alzheimer’s disease. J Neural Transm Suppl 53:147–158
Palop JJ, Mucke L (2010) Amyloid-beta-induced neuronal dysfunction in Alzheimer’s disease: from synapses toward neural networks. Nat Neurosci 13:812–818
Polymeropoulos MH, Lavedan C, Leroy E et al (1997) Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 276:2045–2047
Singleton AB, Farrer MJ, Johnson J et al (2003) alpha-Synuclein locus triplication causes Parkinson’s disease. Science 302:841. doi:10.1126/science.1090278
Simón-Sánchez J, Schulte C, Bras JM et al (2009) Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet 41:1308–1312
Fortin DL, Nemani VM, Voglmaier SM et al (2005) Neural activity controls the synaptic accumulation of alpha-synuclein. J Neurosci 25:10913–10921. doi:10.1523/JNEUROSCI.2922-05.2005
Burré J, Sharma M, Tsetsenis T et al (2010) Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science 329:1663–1667
Chandra S, Gallardo G, Fernández-Chacón R et al (2005) Alpha-synuclein cooperates with CSPalpha in preventing neurodegeneration. Cell 123:383–396
Quilty MC, King AE, Gai W-P et al (2006) Alpha-synuclein is upregulated in neurones in response to chronic oxidative stress and is associated with neuroprotection. Exp Neurol 199:249–256
Nemani VM, Lu W, Berge V et al (2010) Increased expression of alpha-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron 65:66–79
Schulz-Schaeffer WJ (2010) The synaptic pathology of α-synuclein aggregation in dementia with Lewy bodies, Parkinson’s disease and Parkinson’s disease dementia. Acta Neuropathologica 120:131–143
Piccoli G, Condliffe SB, Bauer M et al (2011) LRRK2 controls synaptic vesicle storage and mobilization within the recycling pool. J Neurosci 31:2225–2237
Shin N, Jeong H, Kwon J et al (2008) LRRK2 regulates synaptic vesicle endocytosis. Exp Cell Res 314:2055–2065
Conway KA, Rochet JC, Bieganski RM, Lansbury PT (2001) Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science 294:1346–1349. doi:10.1126/science.1063522
Leong SL, Cappai R, Barnham KJ, Pham CLL (2009) Modulation of alpha-synuclein aggregation by dopamine: a review. Neurochem Res 34:1838–1846
Perier C, Vila M (2012) Mitochondrial biology and Parkinson’s disease. Cold Spring Harb Perspect Med 2:a009332. doi:10.1101/cshperspect.a009332
Tai H-C, Schuman EM (2008) Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat Rev Neurosci 9:826–838
Youle RJ, Van der Bliek AM (2012) Mitochondrial fission, fusion, and stress. Science 337:1062–1065. doi:10.1126/science.1219855
Schapira AHV, Cooper JM, Dexter D et al (1990) Mitochondrial complex I deficiency in Parkinson’s disease. J Neurochem 54:823–827
Parker WD, Boyson SJ, Parks JK (1989) Abnormalities of the electron transport chain in idiopathic Parkinson’s disease. Ann Neurol 26:719–723. doi:10.1002/ana.410260606
Koopman W, Willems P (2012) Monogenic mitochondrial disorders. New Engl J Med
Matsuda N, Sato S, Shiba K et al (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189:211–221. doi:10.1083/jcb.200910140
Plun-Favreau H, Klupsch K, Moisoi N et al (2007) The mitochondrial protease HtrA2 is regulated by Parkinson’s disease-associated kinase PINK1. Nature Cell Biol 9:1243–1252. doi:10.1038/ncb1644
Pridgeon JW, Olzmann J a, Chin L-S, Li L (2007) PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol 5:e172. doi:10.1371/journal.pbio.0050172
Chan NC, Salazar AM, Pham AH et al (2011) Broad activation of the ubiquitin–proteasome system by Parkin is critical for mitophagy. Hum Mol Genet 20:1726–1737. doi:10.1093/hmg/ddr048
Gegg ME, Cooper JM, Chau K-Y et al (2010) Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet 19:4861–4870. doi:10.1093/hmg/ddq419
Geisler S, Holmström KM, Skujat D et al (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature Cell Biol 12:119–131. doi:10.1038/ncb2012
Shin J-H, Ko HS, Kang H et al (2011) PARIS (ZNF746) Repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell 144:689–702. doi:10.1016/j.cell.2011.02.010
Scarpulla RC (2008) Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88:611–638. doi:10.1152/physrev.00025.2007
Ryan MT, Hoogenraad NJ (2007) Mitochondrial-nuclear communications. Annu Rev Biochem 76:701–722. doi:10.1146/annurev.biochem.76.052305.091720
Gandhi S, Wood-Kaczmar A, Yao Z et al (2009) PINK1-associated Parkinson’s disease is caused by neuronal vulnerability to calcium-induced cell death. Molecular Cell 33:627–638. doi:10.1016/j.molcel.2009.02.013
Yao Z, Gandhi S, Burchell VS et al (2011) Cell metabolism affects selective vulnerability in PINK1-associated Parkinson’s disease. J Cell Sci 124:4194–4202. doi:10.1242/jcs.088260
Tschopp J (2011) Mitochondria: Sovereign of inflammation? Eur J Immunol 41:1196–1202
Thomas KJ, McCoy MK, Blackinton J et al (2011) DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet 20:40–50. doi:10.1093/hmg/ddq430
Burbulla LF, Schelling C, Kato H et al (2010) Dissecting the role of the mitochondrial chaperone mortalin in Parkinson’s disease: functional impact of disease-related variants on mitochondrial homeostasis. Hum Mol Genet 19:4437–4452. doi:10.1093/hmg/ddq370
Yang H, Zhou X, Liu X et al (2011) Mitochondrial dysfunction induced by knockdown of mortalin is rescued by Parkin. Biochem Biophys Res Commun 410:114–120. doi:10.1016/j.bbrc.2011.05.116
Weihofen A, Thomas KJ, Ostaszewski BL et al (2009) Pink1 forms a multiprotein complex with Miro and Milton, linking Pink1 function to mitochondrial trafficking. Biochemistry 48:2045–2052. doi:10.1021/bi8019178
Wang X, Winter D, Ashrafi G et al (2011) PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147:893–906. doi:10.1016/j.cell.2011.10.018
Pilsl A, Winklhofer KF (2012) Parkin, PINK1 and mitochondrial integrity: emerging concepts of mitochondrial dysfunction in Parkinson’s disease. Acta Neuropathol 123:173–188. doi:10.1007/s00401-011-0902-3
Sheng Z-H, Cai Q (2012) Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat Rev Neurosci 13:77–93. doi:10.1038/nrn3156
Lee H-J, Khoshaghideh F, Lee S, Lee S-J (2006) Impairment of microtubule-dependent trafficking by overexpression of alpha-synuclein. Eur J Neurosci 24:3153–3162. doi:10.1111/j.1460-9568.2006.05210.x
Yang F, Jiang Q, Zhao J et al (2005) Parkin stabilizes microtubules through strong binding mediated by three independent domains. J Biol Chem 280:17154–17162. doi:10.1074/jbc.M500843200
Gillardon F (2009) Leucine-rich repeat kinase 2 phosphorylates brain tubulin-beta isoforms and modulates microtubule stability—a point of convergence in parkinsonian neurodegeneration? J Neurochem 110:1514–1522. doi:10.1111/j.1471-4159.2009.06235.x
Borland MK, Trimmer PA, Rubinstein JD et al (2008) Chronic, low-dose rotenone reproduces Lewy neurites found in early stages of Parkinson’s disease, reduces mitochondrial movement and slowly kills differentiated SH-SY5Y neural cells. Mol Neurodegener 3:21. doi:10.1186/1750-1326-3-21
Park D-W, Nam M-K, Rhim H (2011) The serine protease HtrA2 cleaves UCH-L1 and inhibits its hydrolase activity: implication in the UCH-L1-mediated cell death. Biochem Biophys Res Commun 415:24–29. doi:10.1016/j.bbrc.2011.09.148
Gusdon AM, Zhu J, Van Houten B, Chu CT (2012) ATP13A2 regulates mitochondrial bioenergetics through macroautophagy. Neurobiol Dis 45:962–972. doi:10.1016/j.nbd.2011.12.015
Braschi E, Goyon V, Zunino R et al (2010) Vps35 mediates vesicle transport between the mitochondria and peroxisomes. Curr Biol 20:1310–1315. doi:10.1016/j.cub.2010.05.066
Chartier-Harlin M-C, Dachsel JC, Vilariño-Güell C et al (2011) Translation initiator EIF4G1 mutations in familial Parkinson disease. Am J Hum Genet 89:398–406. doi:10.1016/j.ajhg.2011.08.009
Lee J, Giordano S, Zhang J (2012) Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J 441:523–540. doi:10.1042/BJ20111451
Korolchuk VI, Menzies FM, Rubinsztein DC (2010) Mechanisms of cross-talk between the ubiquitin–proteasome and autophagy–lysosome systems. FEBS Lett 584:1393–1398
Kroemer G, Mariño G, Levine B (2010) Autophagy and the integrated stress response. Molecular cell 40:280–293. doi:10.1016/j.molcel.2010.09.023
Ravikumar B, Sarkar S, Davies JE et al (2010) Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 90:1383–1435. doi:10.1152/physrev.00030.2009
Harris H, Rubinsztein DC (2011) Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol 8:108–117. doi:10.1038/nrneurol.2011.200
Martinez-Vicente M, Talloczy Z, Kaushik S et al (2008) Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Investig 118:777–788. doi:10.1172/JCI32806DS1
Nalls MA, Plagnol V, Hernandez DG et al (2011) Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet 377:641–649
Grabowski GA (2008) Phenotype, diagnosis, and treatment of Gaucher’s disease. Lancet 372:1263–1271
Yap TL, Gruschus JM, Velayati A et al (2011) {alpha}-Synuclein interacts with glucocerebrosidase providing a molecular link between Parkinson and Gaucher diseases. J Biol Chem 286:28080–28088. doi:10.1074/jbc.M111.237859
Westbroek W, Gustafson AM, Sidransky E (2011) Exploring the link between glucocerebrosidase mutations and parkinsonism. Trends Mol Med 17:485–493
Alvarez-Erviti L, Rodriguez-Oroz MC, Cooper JM et al (2010) Chaperone-mediated autophagy markers in Parkinson disease brains. Arch Neurol 67:1464–1472
Crews L, Spencer B, Desplats P et al (2010) Selective molecular alterations in the autophagy pathway in patients with Lewy body disease and in models of alpha-synucleinopathy. PLoS One 5:e9313
Chu Y, Dodiya H, Aebischer P et al (2009) Alterations in lysosomal and proteasomal markers in Parkinson’s disease: relationship to alpha-synuclein inclusions. Neurobiol Dis 35:385–398. doi:10.1016/j.nbd.2009.05.023
Xu M, Zhang H (2011) Death and survival of neuronal and astrocytic cells in ischemic brain injury: a role of autophagy. Acta Pharmacol Sin 32:1089–1099
Chu CT (2006) Autophagic stress in neuronal injury and disease. J Neuropathol Exp Neurol 65:423–432
Schapira AHV (2012) Targeting mitochondria for neuroprotection in Parkinson’s disease. Antioxid Redox Signal 16:965–973. doi:10.1089/ars.2011.4419
Nixon RA, Yang D-S (2011) Autophagy failure in Alzheimer’s disease—locating the primary defect. Neurobiol Dis 43:38–45. doi:10.1016/j.nbd.2011.01.021
Pickford F, Masliah E, Britschgi M et al (2008) The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Investig 118:2190–2199
Yang D-S, Stavrides P, Mohan PS et al (2011) Therapeutic effects of remediating autophagy failure in a mouse model of Alzheimer disease by enhancing lysosomal proteolysis. Autophagy 7:788–789
Komatsu M, Waguri S, Chiba T et al (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884. doi:10.1038/nature04723
Hara T, Nakamura K, Matsui M et al (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889. doi:10.1038/nature04724
Breydo L, Wu JW, Uversky VN (2012) α-Synuclein misfolding and Parkinson’s disease. Biochim Biophys Acta 1822:261–285. doi:10.1016/j.bbadis.2011.10.002
Ullman O, Fisher CK, Stultz CM (2011) Explaining the structural plasticity of α-synuclein. J Am Chem Soc 133:19536–19546
Wang W, Perovic I, Chittuluru J et al (2011) A soluble α-synuclein construct forms a dynamic tetramer. PNAS 108:17797–17802
Bartels T, Choi JG, Selkoe DJ (2011) α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 3–7. doi: 10.1038/nature10324
Lee S-J, Lim H-S, Masliah E, Lee H-J (2011) Protein aggregate spreading in neurodegenerative diseases: Problems and perspectives. Neurosci Res 70:339–348
Yonetani M, Nonaka T, Masuda M et al (2009) Conversion of wild-type alpha-synuclein into mutant-type fibrils and its propagation in the presence of A30P mutant. J Biol Chem 284:7940–7950
Colla E, Jensen PH, Pletnikova O et al (2012) Accumulation of toxic α-synuclein oligomer within endoplasmic reticulum occurs in α-synucleinopathy in vivo. J Neurosci 32:3301–3305. doi:10.1523/JNEUROSCI.5368-11.2012
Chung CY, Koprich JB, Siddiqi H, Isacson O (2009) Dynamic changes in presynaptic and axonal transport proteins combined with striatal neuroinflammation precede dopaminergic neuronal loss in a rat model of AAV alpha-synucleinopathy. J Neurosci 29:3365–3373. doi:10.1523/JNEUROSCI.5427-08.2009
Ihara M, Yamasaki N, Hagiwara A et al (2007) Sept4, a component of presynaptic scaffold and Lewy bodies, is required for the suppression of alpha-synuclein neurotoxicity. Neuron 53:519–533. doi:10.1016/j.neuron.2007.01.019
Kahle PJ, Neumann M, Ozmen L et al (2000) Subcellular localization of wild-type and Parkinson’s disease-associated mutant alpha-synuclein in human and transgenic mouse brain. J Neurosci 20:6365–6373
Yavich L, Jäkälä P, Tanila H (2006) Abnormal compartmentalization of norepinephrine in mouse dentate gyrus in alpha-synuclein knockout and A30P transgenic mice. J Neurochem 99:724–732. doi:10.1111/j.1471-4159.2006.04098.x
Martin LJ, Pan Y, Price AC et al (2006) Parkinson’s disease alpha-synuclein transgenic mice develop neuronal mitochondrial degeneration and cell death. J Neurosci 26:41–50. doi:10.1523/JNEUROSCI.4308-05.2006
Devi L, Raghavendran V, Prabhu BM et al (2008) Mitochondrial import and accumulation of alpha-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. J Biol Chem 283:9089–9100. doi:10.1074/jbc.M710012200
Loeb V, Yakunin E, Saada A, Sharon R (2010) The transgenic overexpression of alpha-synuclein and not its related pathology associates with complex I inhibition. J Biol Chem 285:7334–7343. doi:10.1074/jbc.M109.061051
Chinta SJ, Mallajosyula JK, Rane A, Andersen JK (2010) Mitochondrial alpha-synuclein accumulation impairs complex I function in dopaminergic neurons and results in increased mitophagy in vivo. Neurosci Lett 486:235–239. doi:10.1016/j.neulet.2010.09.061
Choubey V, Safiulina D, Vaarmann A et al (2011) Mutant A53T alpha-synuclein induces neuronal death by increasing mitochondrial autophagy. J Biol Chem 286:10814–10824. doi:10.1074/jbc.M110.132514
Esposito A, Dohm CP, Kermer P et al (2007) alpha-Synuclein and its disease-related mutants interact differentially with the microtubule protein tau and associate with the actin cytoskeleton. Neurobiol Dis 26:521–531. doi:10.1016/j.nbd.2007.01.014
Lee H-J, Shin SY, Choi C et al (2002) Formation and removal of alpha-synuclein aggregates in cells exposed to mitochondrial inhibitors. J Biol Chem 277:5411–5417. doi:10.1074/jbc.M105326200
Chen L, Jin J, Davis J et al (2007) Oligomeric α-synuclein inhibits tubulin polymerization. Biochem Biophys Res Commun 356:548–553. doi:10.1016/j.bbrc.2007.02.163
Jensen PH (1999) alpha -Synuclein binds to tau and stimulates the protein kinase A-catalyzed tau phosphorylation of serine residues 262 and 356. J Biol Chem 274:25481–25489. doi:10.1074/jbc.274.36.25481
Frasier M, Walzer M, McCarthy L et al (2005) Tau phosphorylation increases in symptomatic mice overexpressing A30P alpha-synuclein. Exp Neurol 192:274–287. doi:10.1016/j.expneurol.2004.07.016
Haggerty T, Credle J, Rodriguez O et al (2011) Hyperphosphorylated Tau in an α-synuclein-overexpressing transgenic model of Parkinson’s disease. Eur J Neurosci 33:1598–1610. doi:10.1111/j.1460-9568.2011.07660.x
Qureshi HY, Paudel HK (2011) Parkinsonian neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and alpha-synuclein mutations promote Tau protein phosphorylation at Ser262 and destabilize microtubule cytoskeleton in vitro. J Biol Chem 286:5055–5068. doi:10.1074/jbc.M110.178905
Clinton LK, Blurton-Jones M, Myczek K et al (2010) Synergistic interactions between Abeta, tau, and alpha-synuclein: acceleration of neuropathology and cognitive decline. J Neurosci 30:7281–7289
Giasson BI, Forman MS, Higuchi M et al (2003) Initiation and synergistic fibrillization of tau and alpha-synuclein. Science 300:636–640. doi:10.1126/science.1082324
Smith WW, Jiang H, Pei Z et al (2005) Endoplasmic reticulum stress and mitochondrial cell death pathways mediate A53T mutant alpha-synuclein-induced toxicity. Hum Mol Genet 14:3801–3811. doi:10.1093/hmg/ddi396
Colla E, Coune P, Liu Y et al (2012) Endoplasmic reticulum stress is important for the manifestations of α-synucleinopathy in vivo. J Neurosci 32:3306–3320
Cuervo AM, Stefanis L, Fredenburg R et al (2004) Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305:1292–1295. doi:10.1126/science.1101738
Xilouri M, Vogiatzi T, Vekrellis K et al (2009) Abberant alpha-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS One 4:e5515. doi:10.1371/journal.pone.0005515
Mazzulli JR, Xu Y-H, Sun Y et al (2011) Gaucher disease glucocerebrosidase and α-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell 146:37–52. doi:10.1016/j.cell.2011.06.001
Tanaka Y, Engelender S, Igarashi S et al (2001) Inducible expression of mutant alpha-synuclein decreases proteasome activity and increases sensitivity to mitochondria-dependent apoptosis. Hum Mol Genet 10:919–926. doi:10.1093/hmg/10.9.919
Stefanis L, Larsen KE, Rideout HJ et al (2001) Expression of A53T mutant but not wild-type alpha-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J Neurosci 21:9549–9560
Petrucelli L, O’Farrell C, Lockhart PJ et al (2002) Parkin protects against the toxicity associated with mutant α-synuclein proteasome dysfunction selectively affects catecholaminergic neurons. Neuron 36:1007–1019. doi:10.1016/S0896-6273(02)01125-X
Lindersson E, Beedholm R, Højrup P et al (2004) Proteasomal inhibition by alpha-synuclein filaments and oligomers. J Biol Chem 279:12924–12934. doi:10.1074/jbc.M306390200
Goedert M, Clavaguera F, Tolnay M (2010) The propagation of prion-like protein inclusions in neurodegenerative diseases. Trends Neurosci 33:317–325
Kordower JH, Chu Y, Hauser RA et al (2008) Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson’s disease. Nature Medicine 14:504–506. doi:10.1038/nm1747
Kordower JH, Chu Y, Hauser RA et al (2008) Transplanted dopaminergic neurons develop PD pathologic changes: a second case report. Mov Disord 23:2303–2306
Li J-Y, Englund E, Holton JL et al (2008) Lewy bodies in grafted neurons in subjects with Parkinson’s disease suggest host-to-graft disease propagation. Nature Medicine 14:501–503. doi:10.1038/nm1746
Desplats P, Lee H-J, Bae E-J et al (2009) Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. PNAS 106:13010–13015. doi:10.1073/pnas.0903691106
Volpicelli-Daley LA, Luk KC, Patel TP et al (2011) Exogenous α-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron 72:57–71
Luk KC, Kehm V, Carroll J et al (2012) Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338:949–953. doi:10.1126/science.1227157
Jang A, Lee H-J, Suk J-E et al (2010) Non-classical exocytosis of alpha-synuclein is sensitive to folding states and promoted under stress conditions. J Neurochem 113:1263–1274
Lee H-J, Patel S, Lee S-J (2005) Intravesicular localization and exocytosis of alpha-synuclein and its aggregates. J Neurosci 25:6016–6024
Luk KC, Song C, O’Brien P et al (2009) Exogenous alpha-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. PNAS 106:20051–20056
Mosley RL, Hutter-Saunders JA, Stone DK, Gendelman HE (2012) Inflammation and adaptive immunity in Parkinson’s disease. Cold Spring Harbor perspectives in medicine 2:a009381.
McGeer PL, Itagaki S, Akiyama H, McGeer EG (1988) Rate of cell death in parkinsonism indicates active neuropathological process. Ann Neurol 24:574–576
Brochard V, Combadière B, Prigent A et al (2009) Infiltration of CD4+ lymphocytes into the brain contributes to neurodegeneration in a mouse model of Parkinson disease. J Clin Investig 119:182–192. doi:10.1172/JCI36470
Double KL, Rowe DB, Carew-Jones FM et al (2009) Anti-melanin antibodies are increased in sera in Parkinson’s disease. Exp Neurol 217:297–301. doi:10.1016/j.expneurol.2009.03.002
Reynolds AD, Stone DK, Hutter J a L et al (2010) Regulatory T cells attenuate Th17 cell-mediated nigrostriatal dopaminergic neurodegeneration in a model of Parkinson’s disease. J Immunol 184:2261–2271. doi:10.4049/jimmunol.0901852, Baltimore, Md: 1950
Lucin KM, Wyss-Coray T (2009) Immune activation in brain aging and neurodegeneration: too much or too little? Neuron 64:110–122
Glass CK, Saijo K, Winner B et al (2010) Mechanisms underlying inflammation in neurodegeneration. Cell 140:918–934. doi:10.1016/j.cell.2010.02.016
Rocha SM, Cristovão AC, Campos FL et al (2012) Astrocyte-derived GDNF is a potent inhibitor of microglial activation. Neurobiol Dis 47:407–415. doi:10.1016/j.nbd.2012.04.014
Nimmerjahn A, Kirchhoff F, Helmchen F (2005) Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308:1314–1318
Davalos D, Grutzendler J, Yang G et al (2005) ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci 8:752–758
Mott RT, Ait-Ghezala G, Town T et al (2004) Neuronal expression of CD22: novel mechanism for inhibiting microglial proinflammatory cytokine production. Glia 46:369–379
Majed HH, Chandran S, Niclou SP et al (2006) A novel role for Sema3A in neuroprotection from injury mediated by activated microglia. J Neurosci 26:1730–1738
Tian L, Rauvala H, Gahmberg CG (2009) Neuronal regulation of immune responses in the central nervous system. Trends Immunol 30:91–99
Koning N, Bö L, Hoek RM, Huitinga I (2007) Downregulation of macrophage inhibitory molecules in multiple sclerosis lesions. Ann Neurol 62:504–514
Cardona AE, Pioro EP, Sasse ME et al (2006) Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci 9:917–924
Kim WG, Mohney RP, Wilson B et al (2000) Regional difference in susceptibility to lipopolysaccharide-induced neurotoxicity in the rat brain: role of microglia. J Neurosci: Off J Soc Neurosci 20:6309–6316
Mena M a, Yébenes G d J (2008) Immune activation in brain aging and neurodegeneration: too much or too little? Neuroscientist 14:544–560. doi:10.1177/1073858408322839
Zecca L, Wilms H, Geick S et al (2008) Human neuromelanin induces neuroinflammation and neurodegeneration in the rat substantia nigra: implications for Parkinson’s disease. Acta Neuropathol 116:47–55. doi:10.1007/s00401-008-0361-7
McGeer PL, Itagaki S, Boyes BE, McGeer EG (1988) Reactive microglia are positive for HLA-DR in the substantia nigra of Parkinson’s and Alzheimer's disease brains. Neurology 38:1285–1285
Langston JW, Forno LS, Tetrud J et al (1999) Evidence of active nerve cell degeneration in the substantia nigra of humans years after 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine exposure. Ann Neurol 46:598–605
McGeer PL, Schwab C, Parent A, Doudet D (2003) Presence of reactive microglia in monkey substantia nigra years after 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine administration. Ann Neurol 54:599–604. doi:10.1002/ana.10728
Czlonkowska A, Kohutnicka M, Kurkowska-Jastrzebska I, Czlonkowski A (1996) Microglial reaction in MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) induced Parkinson’s disease mice model. Neurodegeneration 5:137–143
Walsh S, Finn DP, Dowd E (2011) Time-course of nigrostriatal neurodegeneration and neuroinflammation in the 6-hydroxydopamine-induced axonal and terminal lesion models of Parkinson’s disease in the rat. Neuroscience 175:251–261. doi:10.1016/j.neuroscience.2010.12.005
Gao H, Liu B, Zhang W, Hong J (2003) Critical role of microglial NADPH oxidase-derived free radicals in the in vitro MPTP model of Parkinson’s disease. FASEB J 17:1954–1956. doi:10.1096/fj.03-0109fje
Zhang W, Wang T, Pei Z et al (2005) Aggregated alpha-synuclein activates microglia: a process leading to disease progression in Parkinson’s disease. FASEB J 19:533–542. doi:10.1096/fj.04-2751com
Béraud D, Twomey M, Bloom B et al (2011) α-Synuclein alters Toll-like receptor expression. Frontiers in. Neuroscience 5:80
Zhang W, Dallas S, Zhang D et al (2007) Microglial PHOX and Mac-1 are essential to the enhanced dopaminergic neurodegeneration elicited by A30P and A53T mutant alpha-synuclein. Glia 55:1178–1188
Alvarez-Erviti L, Couch Y, Richardson J et al (2011) Alpha-synuclein release by neurons activates the inflammatory response in a microglial cell line. Neurosci Res 69:337–342. doi:10.1016/j.neures.2010.12.020
Gillardon F, Schmid R, Draheim H (2012) Parkinson’s disease-linked leucine-rich repeat kinase 2(R1441G) mutation increases proinflammatory cytokine release from activated primary microglial cells and resultant neurotoxicity. Neuroscience 208:41–48. doi:10.1016/j.neuroscience.2012.02.001
Frank-Cannon TC, Tran T, Ruhn K a et al (2008) Parkin deficiency increases vulnerability to inflammation-related nigral degeneration. J Neurosci 28:10825–10834. doi:10.1523/JNEUROSCI.3001-08.2008
Waak J, Weber SS, Waldenmaier A et al (2009) Regulation of astrocyte inflammatory responses by the Parkinson’s disease-associated gene DJ-1. FASEB J 23:2478–2489. doi:10.1096/fj.08-125153
Wu DC, Jackson-Lewis V, Vila M et al (2002) Blockade of microglial activation is neuroprotective in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine mouse model of Parkinson disease. J Neurosci 22:1763–1771
Perier C, Bové J, Vila M (2012) Mitochondria and programmed cell death in Parkinson’s disease: apoptosis and beyond. Antioxid Redox Signal 16:883–895. doi:10.1089/ars.2011.4074
Youle RJ, Strasser A (2008) The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 9:47–59. doi:10.1038/nrm2308
Tait SWG, Green DR (2010) Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 11:621–632. doi:10.1038/nrm2952
Offen D, Beart PM, Cheung NS et al (1998) Transgenic mice expressing human Bcl-2 in their neurons are resistant to 6-hydroxydopamine and 1-methyl-4-phenyl-1,2,3,6- tetrahydropyridine neurotoxicity. PNAS 95:5789–5794
Yang L, Matthews RT, Schulz JB et al (1998) 1-Methyl-4-phenyl-1,2,3,6-tetrahydropyride neurotoxicity is attenuated in mice overexpressing Bcl-2. J Neurosci 18:8145–8152
Vila M, Jackson-Lewis V, Vukosavic S et al (2001) Bax ablation prevents dopaminergic neurodegeneration in the 1-methyl- 4-phenyl-1,2,3,6-tetrahydropyridine mouse model of Parkinson’s disease. PNAS 98:2837–2842. doi:10.1073/pnas.051633998
Perier C, Bové J, Wu D-C et al (2007) Two molecular pathways initiate mitochondria-dependent dopaminergic neurodegeneration in experimental Parkinson’s disease. PNAS 104:8161–8166. doi:10.1073/pnas.0609874104
Dietz GPH, Stockhausen KV, Dietz B et al (2008) Membrane-permeable Bcl-xL prevents MPTP-induced dopaminergic neuronal loss in the substantia nigra. J Neurochem 104:757–765. doi:10.1111/j.1471-4159.2007.05028.x
Iaccarino C, Crosio C, Vitale C et al (2007) Apoptotic mechanisms in mutant LRRK2-mediated cell death. Hum Mol Genet 16:1319–1326. doi:10.1093/hmg/ddm080
Darios F, Corti O, Lücking CB et al (2003) Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum Mol Genet 12:517–526
Petit A, Kawarai T, Paitel E et al (2005) Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J Biol Chem 280:34025–34032. doi:10.1074/jbc.M505143200
Wang H-L, Chou A-H, Yeh T-H et al (2007) PINK1 mutants associated with recessive Parkinson’s disease are defective in inhibiting mitochondrial release of cytochrome c. Neurobiol Dis 28:216–226. doi:10.1016/j.nbd.2007.07.010
Boya P, Kroemer G (2008) Lysosomal membrane permeabilization in cell death. Oncogene 27:6434–6451. doi:10.1038/onc.2008.310
Turk B, Turk DSA, Turk V (2012) Protease signalling: the cutting edge. EMBO J 31:1630–1643. doi:10.1038/emboj.2012.42
Jeon S-M, Cheon S-M, Bae H-R et al (2010) Selective susceptibility of human dopaminergic neural stem cells to dopamine-induced apoptosis. Exp Neurobiol 19:155–164. doi:10.5607/en.2010.19.3.155
Benner EJ, Banerjee R, Reynolds AD et al (2008) Nitrated alpha-synuclein immunity accelerates degeneration of nigral dopaminergic neurons. PLoS One 3:e1376. doi:10.1371/journal.pone.0001376
Gao H-M, Zhou H, Zhang F et al (2011) HMGB1 acts on microglia Mac1 to mediate chronic neuroinflammation that drives progressive neurodegeneration. J Neurosci 31:1081–1092. doi:10.1523/JNEUROSCI.3732-10.2011
Simi A, Tsakiri N, Wang P, Rothwell NJ (2007) Interleukin-1 and inflammatory neurodegeneration. Biochem Soc Trans 35:1122–1126
McCoy MK, Tansey MG (2008) TNF signaling inhibition in the CNS: implications for normal brain function and neurodegenerative disease. J Neuroinflammation 5:45. doi:10.1186/1742-2094-5-45
Magrane M, Consortium U (2011) UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011:bar009.
Seal RL, Gordon SM, Lush MJ et al (2011) genenames.org: the HGNC resources in 2011. Nucleic Acids Res 39:D514–D519
Cochrane G, Akhtar R, Bonfield J et al (2009) Petabyte-scale innovations at the European Nucleotide Archive. Nucleic Acids Res 37:19–25
Flicek P, Amode MR, Barrell D et al (2012) Ensembl 2012. Nucleic Acids Res 40:84–90
Fujita PA, Rhead B, Zweig AS et al (2011) The UCSC Genome Browser database: update 2011. Nucleic Acids Res 39:D876–D882
Pruitt KD, Tatusova T, Brown GR, Maglott DR (2012) NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res 40:D130–D135
Maglott D, Ostell J, Pruitt KD, Tatusova T (2011) Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 39:D52–D57
Schuler GD (1997) Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J Mol Med 75:694–698
Safran M, Dalah I, Alexander J, et al. (2010) Gene Cards Version 3: the human gene integrator. Database (Oxford) 2010:baq020.
Berman H, Henrick K, Nakamura H, Markley JL (2007) The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res 35:D301–D303
Ashburner M, Ball CA, Blake JA et al (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29
Kanehisa M, Goto S, Sato Y et al (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40:D109–D114
Mi H, Lazareva-Ulitsky B, Loo R et al (2005) The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res 33:D284–D288
Croft D, O’Kelly G, Wu G et al (2011) Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res 39:D691–D697
Punta M, Coggill PC, Eberhardt RY et al (2012) The Pfam protein families database. Nucleic Acids Res 40:290–301
Hunter S, Jones P, Mitchell A et al (2012) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res 40:D306–D312
cDonagh EM, Whirl-Carrillo M, Garten Y et al (2011) From pharmacogenomic knowledge acquisition to clinical applications: the PharmGKB as a clinical pharmacogenomic biomarker resource. Biomark Med 5:795–806
Zhang Y, James M, Middleton FA, Davis RL (2005) Transcriptional analysis of multiple brain regions in Parkinson’s disease supports the involvement of specific protein processing, energy metabolism, and signaling pathways, and suggests novel disease mechanisms. Am J Med Genet 137B:5–16. doi:10.1002/ajmg.b.30195
Lesnick TG, Papapetropoulos S, Mash DC et al (2007) A genomic pathway approach to a complex disease: axon guidance and Parkinson disease. PLoS Genet 3:12
Moran LB, Duke DC, Deprez M et al (2006) Whole genome expression profiling of the medial and lateral substantia nigra in Parkinson’s disease. Neurogenetics 7:1–11
Zheng B, Liao Z, Locascio JJ et al (2010) PGC-1α, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med 2:52ra–73ra
Scherzer CR, Eklund AC, Morse LJ et al (2007) Molecular markers of early Parkinson’s disease based on gene expression in blood. PNAS 104:955–960
Sforza D, Hartenstein P, Lacan G, et al. (2008) Gene expression changes in multiple brain regions of a mouse MPTP model of Parkinson’s disease. Fairfax, VA 22030 USA
Foti R, Zucchelli S, Biagioli M et al (2010) Parkinson disease-associated DJ-1 is required for the expression of the glial cell line-derived neurotrophic factor receptor RET in human neuroblastoma cells. J Biol Chem 285:18565–18574
Rajagopalan D, Agarwal P (2005) Inferring pathways from gene lists using a literature-derived network of biological relationships. Bioinformatics 21:788–793. doi:10.1093/bioinformatics/bti069
Ulitsky I, Shamir R (2009) Identifying functional modules using expression profiles and confidence-scored protein interactions. Bioinformatics 25:1158–1164. doi:10.1093/bioinformatics/btp118
Cerami E, Demir E, Schultz N et al (2010) Automated network analysis identifies core pathways in glioblastoma. PLoS One 5:e8918. doi:10.1371/journal.pone.0008918
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. PNAS 98:5116–5121. doi:10.1073/pnas.091062498
Aerts S, Lambrechts D, Maity S et al (2006) Gene prioritization through genomic data fusion. Nat Biotechnol 24:537–544. doi:10.1038/nbt1203
Ma X, Lee H, Wang L, Sun F (2007) CGI: a new approach for prioritizing genes by combining gene expression and protein–protein interaction data. Bioinformatics 23:215–221. doi:10.1093/bioinformatics/btl569
Zaykin DV, Zhivotovsky LA, Czika W et al (2007) Combining p-values in large-scale genomics experiments. Pharm Stat 6:217–226. doi:10.1002/pst.304
Marot G, Foulley J-L, Mayer C-D, Jaffrézic F (2009) Moderated effect size and P-value combinations for microarray meta-analyses. Bioinformatics 25:2692–2699
Aittokallio T, Schwikowski B (2006) Graph-based methods for analysing networks in cell biology. Brief Bioinform 7:243–255. doi:10.1093/bib/bbl022
Emmert-Streib F, Dehmer M (2011) Networks for systems biology: conceptual connection of data and function. IET Syst Biol 5:185. doi:10.1049/iet-syb.2010.0025
Junker BH, Schreiber F (2008) Analysis of biological networks. Science 1–28. doi:10.1002/9780470253489
Guimerà R, Sales-Pardo M, Amaral LAN (2007) Classes of complex networks defined by role-to-role connectivity profiles. Nat Phys 3:63–69. doi:10.1038/nphys489
Wang R-S, Albert R (2011) Elementary signaling modes predict the essentiality of signal transduction network components. BMC Syst Biol 5:44
Klipp E, Liebermeister W (2006) Mathematical modeling of intracellular signaling pathways. BMC Neurosci 7(Suppl 1):S10. doi:10.1186/1471-2202-7-S1-S10
Hucka M, Finney a, Sauro HM (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19:524–531. doi:10.1093/bioinformatics/btg015
Hoops S, Sahle S, Gauges R et al (2006) COPASI—a COmplex PAthway SImulator. Bioinform (Oxford, England) 22:3067–3074. doi:10.1093/bioinformatics/btl485
Van Eunen K, Kiewiet J a L, Westerhoff HV, Bakker BM (2012) Testing biochemistry revisited: how in vivo metabolism can be understood from in vitro enzyme kinetics. PLoS Comput Biol 8:e1002483. doi:10.1371/journal.pcbi.1002483
Kowald A, Hamann A, Zintel S et al (2012) A systems biological analysis links ROS metabolism to mitochondrial protein quality control. MechAgeing Dev 133:331–337. doi:10.1016/j.mad.2012.03.008
Berndt N, Bulik S, Holzhütter H-G (2012) Kinetic modeling of the mitochondrial energy metabolism of neuronal cells: the impact of reduced α-ketoglutarate dehydrogenase activities on ATP production and generation of reactive oxygen species. International journal of cell biology 2012:757594. doi: 10.1155/2012/757594
Craddock TJ a, Tuszynski J a, Chopra D et al (2012) The zinc dyshomeostasis hypothesis of Alzheimer’s disease. PLoS One 7:e33552. doi:10.1371/journal.pone.0033552
Ambert N, Greget R, Haeberlé O et al (2010) Computational studies of NMDA receptors: differential effects of neuronal activity on efficacy of competitive and non-competitive antagonists. Open Access Bioinforma 2:113–125. doi:10.2147/OAB.S7246
Berardini TZ, Li D, Muller R, et al. (2012) Assessment of community-submitted ontology annotations from a novel database-journal partnership. Database : the journal of biological databases and curation 2012:bas030. doi: 10.1093/database/bas030
Meyer P, Alexopoulos LG, Bonk T et al (2011) Verification of systems biology research in the age of collaborative competition. Nat Biotechnol 29:811–815. doi:10.1038/nbt.1968
Meyer P, Hoeng J, Rice JJ et al (2012) Industrial methodology for process verification in research (IMPROVER): toward systems biology verification. Bioinform (Oxford, England) 28:1193–1201. doi:10.1093/bioinformatics/bts116
Nielsen MA (2011) Reinventing discovery: the new era of networked science. Princeton University Press, Princeton, NJ
Kelder T, Van Iersel MP, Hanspers K et al (2012) WikiPathways: building research communities on biological pathways. Nucleic Acids Res 40:D1301–D1307. doi:10.1093/nar/gkr1074
Matsuoka Y, Ghosh S, Kikuchi N, Kitano H (2010) Payao: a community platform for SBML pathway model curation. Bioinformatics 26:1381–1383. doi:10.1093/bioinformatics/btq143
Le Novère N, Hucka M, Mi H et al (2009) The systems biology graphical notation. Nat Biotechnol 27:735–741
Acknowledgments
We are grateful to Angela Hogan for language correction assistance.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
K. Fujita and M. Ostaszewski contributed equally to this work.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Fujita, K.A., Ostaszewski, M., Matsuoka, Y. et al. Integrating Pathways of Parkinson's Disease in a Molecular Interaction Map. Mol Neurobiol 49, 88–102 (2014). https://doi.org/10.1007/s12035-013-8489-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-013-8489-4