Abstract
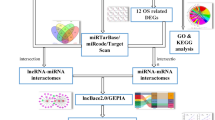
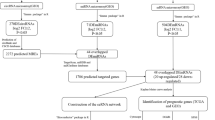
In the fight against glioblastoma, circular RNA is emerging as a functional molecule. However, how circular RNA (circRNA) is regulated and what role it plays is still a mystery. In this research, different bioinformatics approaches were used to evaluate glioblastoma circRNA sequencing and array data, with the goal of developing a putative molecular sponge mechanism control network. The circRNAs were obtained from the Gene Expression Omnibus datasets. MicroRNA-circRNA interactions were predicted using CircInteractome. The microRNAs' expression and survival trends were screened using the TCGA database. MicroRNA gene targets were predicted using the MiRnet database. Sponge network gene candidates were screened using data from the GEPIA. The roles of the targeted genes were to be explained by analyzing data from Gene Ontology and the Kyoto Encyclopedia of Genes and Genomes. To build the network and display the outcomes, we utilized python program, and enrichment online Bioinformatics databases. The circRNAs hsa_circITGA4_002, hsa_circITGA4_001, hsa_circITGA4_003, hsa_circ_0030855, hsa_circ_0030857 were chosen from among GBM patients and control group. Upregulation of hsa-miR-1468, hsa-miR-3683, hsa-miR-1273c, and hsa-miR-4665-3p were associated with a poor prognosis in GBM. MicroRNA targets such as ITGA4, LAMA2, EGFR, PTEN, COL1A4, and NCAM2 were analyzed using expression and survival data. The Apoptosis, cell adhesion molecules, PI3K/AKT and P53 signaling pathways were the most abundant functional categories among gene targets. The circRNA molecular sponge regulatory network includes hsa-miR-1468 and hsa-miR-4665-3p. In this network, hs hsa_circITGA4_002, hsa_circITGA4_001, hsa_circ_0030857, EGFR, PTEN, and ITGA4 may represent GBM therapeutic targets. Their role in GBM needs additional study.






Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article.
References
Kaptein, F. H. J., Stals, M. A. M., Kapteijn, M. Y., Cannegieter, S. C., Dirven, L., van Duinen, S. G., et al. (2022). Incidence and determinants of thrombotic and bleeding complications in patients with glioblastoma. Journal of Thrombosis and Haemostasis, 20(7), 1665–1673.
Tan, A. C., Ashley, D. M., López, G. Y., Malinzak, M., Friedman, H. S., & Khasraw, M. (2020). Management of glioblastoma: State of the art and future directions. CA: A Cancer Journal for Clinicians, 70(4), 299–312.
Fan, X., Xiong, Y., & Wang, Y. (2019). A reignited debate over the cell(s) of origin for glioblastoma and its clinical implications. Frontiers in Medicine, 13(5), 531–539.
AlcantaraLlaguno, S., Sun, D., Pedraza, A. M., Vera, E., Wang, Z., Burns, D. K., et al. (2019). Cell-of-origin susceptibility to glioblastoma formation declines with neural lineage restriction. Nature Neuroscience, 22(4), 545–555.
Wen, P. Y., Weller, M., Lee, E. Q., Alexander, B. M., Barnholtz-Sloan, J. S., Barthel, F. P., et al. (2020). Glioblastoma in adults: A Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro-Oncology, 22(8), 1073–1113.
Bahadur, S., Sahu, A. K., Baghel, P., & Saha, S. (2019). Current promising treatment strategy for glioblastoma multiform: A review. Oncology Reviews, 13(2), 417.
Sanders, S., & Debinski, W. (2020). Challenges to successful implementation of the immune checkpoint inhibitors for treatment of glioblastoma. International Journal of Molecular Sciences, 21(8), 2759.
Majc, B., Novak, M., Kopitar-Jerala, N., Jewett, A., & Breznik, B. (2021). Immunotherapy of glioblastoma: Current strategies and challenges in tumor model development. Cells, 10(2), 265.
Wang, J., Zhu, S., Meng, N., He, Y., Lu, R., & Yan, G.-R. (2019). ncRNA-encoded peptides or proteins and cancer. Molecular Therapy, 27(10), 1718–1725.
Fabbri, M., Girnita, L., Varani, G., & Calin, G. A. (2019). Decrypting noncoding RNA interactions, structures, and functional networks. Genome Research, 29(9), 1377–1388.
Shi, Y., Jia, X., & Xu, J. (2020). The new function of circRNA: Translation. Clinical and Translational Oncology, 22(12), 2162–2169.
Cao, Y.-Z., Sun, J.-Y., Chen, Y.-X., Wen, C.-C., & Wei, L. (2021). The roles of circRNAs in cancers: Perspectives from molecular functions. Gene, 767, 145182.
Hosseini, M.M., Karimi, A., Behroozaghdam, M., Javidi, M.A., Ghiasvand, S., Bereimipour, A., et al. (2017). Cytotoxic and apoptogenic effects of cyanidin-3-glucoside on the glioblastoma cell line. World Neurosurgery, 108, 94–100.
Chen, J., Gu, J., Tang, M., Liao, Z., Tang, R., Zhou, L., et al. (2022). Regulation of cancer progression by circRNA and functional proteins. Journal of Cellular Physiology, 237(1), 373–388.
Liu, Y., Chen, G., Wang, B., Wu, H., Zhang, Y., & Ye, H. (2021). Silencing circRNA protein kinase C iota (circ-PRKCI) suppresses cell progression and glycolysis of human papillary thyroid cancer through circ-PRKCI/miR-335/E2F3 ceRNA axis. Endocrine Journal., 68(6), 713–727.
Hong, W., Xue, M., Jiang, J., Zhang, Y., & Gao, X. (2020). Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). Journal of Experimental & Clinical Cancer Research., 39(1), 1–19.
Liu, J., Hou, K., Ji, H., Mi, S., Yu, G., Hu, S., et al. (2020). Overexpression of circular RNA circ-CDC45 facilitates glioma cell progression by sponging miR-516b and miR-527 and predicts an adverse prognosis. Journal of Cellular Biochemistry, 121(1), 690–697.
Jin, Y., Li, L., Zhu, T., & Liu, G. (2019). Circular RNA circ\_0102049 promotes cell progression as ceRNA to target MDM2 via sponging miR-1304-5p in osteosarcoma. Pathology Practice, 215(12), 152688.
Guo, Q., Guo, J., Liu, W., Hu, S., Hu, X., Wang, Q., et al. (2022). Circ-EGFR functions as an inhibitory factor in the malignant progression of glioma by regulating the miR-183-5p/TUSC2 axis. Cellular and Molecular Neurobiology, 42(7), 2245–2256.
Zhang, Y., Geng, X., Xu, J., Li, Q., Hao, L., Zeng, Z., et al. (2021). Identification and characterization of N6-methyladenosine modification of circRNAs in glioblastoma. Journal of Cellular and Molecular Medicine, 25(15), 7204–7217.
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M., & Ishiguro-Watanabe, M. (2023). KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Research, 51(D1), D587–D592.
Chen, L., & Shan, G. (2021). CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Letters, 505, 49–57.
Kristensen, L. S., Jakobsen, T., Hager, H., & Kjems, J. (2022). The emerging roles of circRNAs in cancer and oncology. Nature Reviews Clinical Oncology, 19(3), 188–206.
Arnaiz, E., Sole, C., Manterola, L., Iparraguirre, L., Otaegui, D., & Lawrie, C. H. (2019). CircRNAs and cancer: Biomarkers and master regulators. In T. Vincent (Ed.), Seminars in cancer biology (pp. 90–99). London: Academic Press.
Visci, G., Tolomeo, D., Agostini, A., Traversa, D., Macchia, G., & Storlazzi, C. T. (2020). CircRNAs and fusion-circRNAs in cancer: New players in an old game. Cellular Signalling, 75, 109747.
Liu, X., Wen, J., Li, C., Wang, H., Wang, J., & Zou, H. (2020). High-yield methylation markers for stool-based detection of colorectal cancer. Digestive Diseases and Sciences, 65(6), 1710–1719.
Mo, J., Zhang, J., Huang, H., Liu, C., Cheng, Y., Mo, Y., et al. (2022). The early predictive effect of low expression of the ITGA4 in colorectal cancer. Journal of Gastrointestinal Oncology, 13(1), 265.
Fang, T., Yin, X., Wang, Y., Wang, H., Wang, X., Xue, Y., et al. (2022). Lymph node metastasis-related gene ITGA4 promotes the proliferation, migration, and invasion of gastric cancer cells by regulating tumor immune microenvironment. Journal of Oncology, 2022, 1315677.
Long, T., Li, X., Zhang, G., Qiu, C., Huan, O., Sun, C., et al. (2021). Single nucleotide polymorphism mutation related genes in bladder cancer for the treatment of patients: A study based on the TCGA database. Biotechnology & Biotechnological Equipment, 35(1), 214–223.
Li, S., Hu, J., Li, G., Mai, H., Gao, Y., Liang, B., et al. (2022). Epigenetic regulation of LINC01270 in breast cancer progression by mediating LAMA2 promoter methylation and MAPK signaling pathway. Cell Biology and Toxicology. https://doi.org/10.1007/s10565-022-09763-9
Uribe, M. L., Marrocco, I., & Yarden, Y. (2021). EGFR in cancer: Signaling mechanisms, drugs, and acquired resistance. Cancers (Basel), 13(11), 2748.
She, K., Fang, S., Du, W., Fan, X., He, J., Pan, H., et al. (2019). SCD1 is required for EGFR-targeting cancer therapy of lung cancer via re-activation of EGFR/PI3K/AKT signals. Cancer Cell International, 19(1), 1–11.
Duwa, R., Banstola, A., Emami, F., Jeong, J.-H., Lee, S., & Yook, S. (2020). Cetuximab conjugated temozolomide-loaded poly (lactic-co-glycolic acid) nanoparticles for targeted nanomedicine in EGFR overexpressing cancer cells. Journal of Drug Delivery Science and Technology, 60, 101928.
Soto-Gamez, A., Chen, D., Nabuurs, A. G. E., Quax, W. J., Demaria, M., & Boersma, Y. L. (2020). A bispecific inhibitor of the EGFR/ADAM17 axis decreases cell proliferation and migration of EGFR-dependent cancer cells. Cancers (Basel), 12(2), 411.
Raoof, S., Mulford, I. J., Frisco-Cabanos, H., Nangia, V., Timonina, D., Labrot, E., et al. (2019). Targeting FGFR overcomes EMT-mediated resistance in EGFR mutant non-small cell lung cancer. Oncogene, 38(37), 6399–6413.
Acknowledgements
We thank all colleagues in the core facilities for their support.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors Participated in study design, data collection and evaluation, drafting and statistical analysis. Contributed extensively in interpretation of the data and the conclusion and figure design. All authors performed editing and approving the final version of this paper for submission, also participated in the finalization of the manuscript and approved the final draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tutunchi, S., Bereimipour, A. & Ghaderian, S.M.H. Hsa_circITGA4/ miR-1468/EGFR/ PTEN a Master Regulators Axis in Glioblastoma Development and Progression. Mol Biotechnol 66, 90–101 (2024). https://doi.org/10.1007/s12033-023-00735-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-023-00735-w