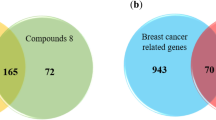
Abstract
Triple-negative breast cancer (TNBC), is diagnosed as the most lethal molecular subtype of breast cancer (BC) preceded by an extremely poor prognosis. For enabling effective TNBC therapy, the identification of novel druggable biomarkers is an earnest need. Multigene paneling and genomewide association studies identify multiple genes with high-to-moderate penetrance in TNBC. Modern computer-aided drug designing techniques, thus aim to design more cost-effective natural small molecule inhibitors for TNBC prevention and diagnosis. Here Amygdalin, a natural glycosidic inhibitor is docked and simulated against three such high-to-moderate penetrance genes identified in TNBC, BARD1, RAD51, and PALB2. The preliminary result of the analysis, reports a highest, intermediate, and least binding energy score of − 6.69 kcal/mol, − 5.09 kcal/mol, and − 4.89 kcal/mol in BARD1, RAD51, and PALB2, respectively. The best-docked protein–ligand complex (BARD1-Amygdalin) was then simulated and compared with an approved drug for TNBC treatment, Olaparib. A comparable binding energy score of − 8.53 kcal/mol was obtained by docking olaparib with BARD1. A 100 ns MD simulation revealed, Amygdalin forms more H-bonds, providing more stable and compact protein–ligand complex with BARD1 than compared to Olaparib. The result was also supported by calculation of solvent accessible surface area and analysis of radius of gyration. Thus, our findings suggest that role of Amygdalin can further be studied in details for TNBC therapeutics, which was found to target the BRCT domain of the BARD1 receptor in stable manner. Please check and confirm that the authors and their respective affiliations have been correctly identified and amend if necessary. Name and affiliations are correctly identified.














Similar content being viewed by others
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
References
Romano, J. D., & Tatonetti, N. P. (2019). Informatics and computational methods in natural product drug discovery: A review and perspectives. Frontiers in genetics, 10, 368.
Mitra, S., & Dash, R. (2018). Natural products for the management and prevention of breast cancer. Evidence-Based Complementary and Alternative Medicine. https://doi.org/10.1155/2018/8324696
Ashburn, T. T., & Thor, K. B. (2004). Drug repositioning: Identifying and developing new uses for existing drugs. Nature reviews Drug discovery, 3(8), 673–683.
Ke, Y. Y., Singh, V. K., Coumar, M. S., Hsu, Y. C., Wang, W. C., Song, J. S., Chen, C. H., Lin, W. H., Wu, S. H., Hsu, J. T., & Shih, C. (2015). Homology modeling of DFG-in FMS-like tyrosine kinase 3 (FLT3) and structure-based virtual screening for inhibitor identification. Scientific Reports, 5(1), 1–12.
Schaduangrat, N., Lampa, S., Simeon, S., Gleeson, M. P., Spjuth, O., & Nantasenamat, C. (2020). Towards reproducible computational drug discovery. Journal of Cheminformatics, 12, 1–30.
Pagadala, N. S., Syed, K., & Tuszynski, J. (2017). Software for molecular docking: A review. Biophysical reviews, 9(2), 91–102.
Salmaso, V., & Moro, S. (2018). Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: an overview. Frontiers Pharmacology. https://doi.org/10.3389/fphar.2018.00923
Dutta, S., Kharkar, P. S., Sahu, N. U., & Khanna, A. (2017). Molecular docking prediction and in vitro studies elucidate anti-cancer activity of phytoestrogens. Life sciences, 185, 73–84.
Ferreira, L. G., Dos Santos, R. N., Oliva, G., & Andricopulo, A. D. (2015). Molecular docking and structure-based drug design strategies. Molecules, 20(7), 13384–13421.
Shi, J., Chen, Q., Xu, M., Xia, Q., Zheng, T., Teng, J., Li, M., & Fan, L. (2019). Recent updates and future perspectives about amygdalin as a potential anticancer agent: a review. Cancer Medicine, 8(6), 3004–3011.
Erkan ÖN, GHOSH A, GUNGOR M, YALIN AE, YALIN S. (2021) “Investigation of molecular modeling and molecular dynamics simulation In BRCA-1 And BRCA-2 genes of amygdalin ligand.”
Jiagang, D., Li, C., Wang, H., Hao, E., Du, Z., Bao, C., Lv, J., & Wang, Y. (2011). Amygdalin mediates relieved atherosclerosis in apolipoprotein E deficient mice through the induction of regulatory T cells. Biochemical and Biophysical Research Communications, 411(3), 523–529.
Orlikova, B., Legrand, N., Panning, J., Dicato, M., & Diederich, M. (2014). Anti-inflammatory and anticancer drugs from nature. Advances in Nutrition and Cancer. https://doi.org/10.1007/978-3-642-38007-5_8
Lea, M. A., & Koch, M. R. (1979). Effects of cyanate, thiocyanate, and amygdalin on metabolite uptake in normal and neoplastic tissues of the rat. Journal of the National Cancer Institute, 63(5), 1279–1283.
Qian, L., Xie, B., Wang, Y., & Qian, J. (2015). Amygdalin-mediated inhibition of non-small cell lung cancer cell invasion in vitro. International journal of clinical and experimental pathology, 8(5), 5363.
Makarević, J., Rutz, J., Juengel, E., Kaulfuss, S., Reiter, M., Tsaur, I., Bartsch, G., Haferkamp, A., & Blaheta, R. A. (2014). Amygdalin blocks bladder cancer cell growth in vitro by diminishing cyclin A and cdk2. PLoS ONE, 9(8), e105590.
Abboud, M. M., Al Awaida, W., Alkhateeb, H. H., & Abu-Ayyad, A. N. (2019). Antitumor action of amygdalin on human breast cancer cells by selective sensitization to oxidative stress. Nutrition and Cancer., 71(3), 483–490.
Mani, J., Neuschäfer, J., Resch, C., Rutz, J., Maxeiner, S., Roos, F., Chun, F. K., Juengel, E., & Blaheta, R. A. (2020). Amygdalin modulates prostate cancer cell adhesion and migration in vitro. Nutrition and Cancer, 72(3), 528–537.
Song, Z., & Xiaohong, Xu. (2014). Advanced research on anti-tumor effects of amygdalin. Journal of Cancer Research and Therapeutics, 10(5), 3.
Zhao, B., Xu, Y., Zhao, Y., Shen, S., & Sun, Q. (2020). Identification of potential key genes associated with the pathogenesis, metastasis, and prognosis of triple-negative breast cancer on the basis of integrated bioinformatics analysis. Frontiers in Oncology. https://doi.org/10.3389/fonc.2020.00856
Zagami, P., & Carey, L. A. (2022). Triple negative breast cancer: Pitfalls and progress. NPJ Breast Cancer, 8(1), 1–10.
Lehmann, B. D., Bauer, J. A., Chen, X., Sanders, M. E., Chakravarthy, A. B., Shyr, Y., & Pietenpol, J. A. (2011). Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of Clinical Investigation, 121(7), 2750–2767.
Montagna, E., Maisonneuve, P., Rotmensz, N., Cancello, G., Iorfida, M., Balduzzi, A., Galimberti, V., Veronesi, P., Luini, A., Pruneri, G., & Bottiglieri, L. (2013). Heterogeneity of triple-negative breast cancer: histologic subtyping to inform the outcome. Clinical Breast Cancer, 13(1), 31–39.
Ellsworth, D. L., Turner, C. E., & Ellsworth, R. E. (2019). A review of the hereditary component of triple negative breast cancer: High-and moderate-penetrance breast cancer genes, low-penetrance loci, and the role of nontraditional genetic elements. Journal of Oncology. https://doi.org/10.1155/2019/4382606
Rebbeck, T. R., Mitra, N., Wan, F., Sinilnikova, O. M., Healey, S., McGuffog, L., Mazoyer, S., Chenevix-Trench, G., Easton, D. F., Antoniou, A. C., & Nathanson, K. L. (2015). Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA, 313(13), 1347–1361.
Brigham & Women’s Hospital & Harvard Medical School Chin Lynda 9 11 Park Peter J. 12 Kucherlapati Raju 13, et al. (2012) “Comprehensive molecular portraits of human breast tumours”. Nature 490(7418): 61–70.
Borri, Filippo and Annarita Granaglia 2021 Pathology of triple negative breast cancer, Seminars in cancer biology, Academic Press
Fornier, M., & Fumoleau, P. (2012). The paradox of triple negative breast cancer: Novel approaches to treatment. The Breast Journal, 18(1), 41–51.
Wahba, H. A., & El-Hadaad, H. A. (2015). Current approaches in treatment of triple-negative breast cancer. Cancer Biology & Medicine, 12(2), 106.
Abbasi, B. A., Iqbal, J., Mahmood, T., Khalil, A. T., Ali, B., Kanwal, S., Shah, S. A., & Ahmad, R. (2018). Role of dietary phytochemicals in modulation of miRNA expression: Natural swords combating breast cancer. Asian Pacific Journal of Tropical Medicine, 11(9), 501.
Rahman, N., Seal, S., Thompson, D., Kelly, P., Renwick, A., Elliott, A., Reid, S., Spanova, K., Barfoot, R., Chagtai, T., & Jayatilake, H. (2007). PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nature Genetics, 39(2), 165–167.
Han, M. R., Zheng, W., Cai, Q., Gao, Y. T., Zheng, Y., Bolla, M. K., Michailidou, K., Dennis, J., Wang, Q., Dunning, A. M., & Brennan, P. (2017). Evaluating genetic variants associated with breast cancer risk in high and moderate-penetrance genes in Asians. Carcinogenesis, 38(5), 511–518.
Oliver, A. W., Swift, S., Lord, C. J., Ashworth, A., & Pearl, L. H. (2009). Structural basis for recruitment of BRCA2 by PALB2. EMBO Reports, 10(9), 990–996.
Xu, J., Zhao, L., Xu, Y., Zhao, W., Sung, P., & Wang, H. W. (2017). Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange. Nature Structural & Molecular Biology, 24(1), 40–46.
Birrane, G., Varma, A. K., Soni, A., & Ladias, J. A. (2007). Crystal structure of the BARD1 BRCT domains. Biochemistry, 46(26), 7706–7712.
Feng, H. B. J. W. Z., Weissig, G. G. T. B. H., Shindyalov, I. N., & Bourne, P. E. (2000). IN Shindyalov PE Bourne. Nucleic Acids Research, 28, 235–242.
Studio, Discovery. “Discovery studio”. Accelrys [4.5] (2021).
Tai, W., He, L., Zhang, X., Pu, J., Voronin, D., Jiang, S., Zhou, Y., & Du, L. (2020). Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cellular & Molecular Immunology, 17(6), 613–620.
Binkowski, T., Andrew, S. N., & Liang, J. (2003). CASTp: computed atlas of surface topography of proteins. Nucleic acids research, 31(13), 3352–3355.
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of computational Chemistry., 30(16), 2785–2791.
Trott, O., and A. J. Olson. (2009) “Software news and update AutoDock Vina: improving the speed and accuracy of docking with a new scoring function”. Efficient Optimization, and Multithreading.
Kim, G., Ison, G., McKee, A. E., Zhang, H., Tang, S., Gwise, T., Sridhara, R., Lee, E., Tzou, A., Philip, R., & Chiu, H. J. (2015). FDA approval summary: Olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapyolaparib for advanced ovarian cancer with BRCA mutation. Clinical Cancer Research, 21(19), 4257–4261.
Laskowski, Roman A., and Mark B. Swindells. (2011) “LigPlot+: multiple ligand–protein interaction diagrams for drug discovery”: 2778–2786.
Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., & Berendsen, H. J. (2005). GROMACS: fast, flexible, and free. Journal of computational chemistry., 26(16), 1701–1718.
Stroet, M., Caron, B., Visscher, K. M., Geerke, D. P., Malde, A. K., & Mark, A. E. (2018). Automated topology builder version 3.0: Prediction of solvation free enthalpies in water and hexane. Journal of Chemical Theory and Computation, 14(11), 5834–5845.
Izadi, S., Anandakrishnan, R., & Onufriev, A. V. (2014). Building water models: A different approach. The journal of physical chemistry letters, 5(21), 3863–3871.
Ross, G. A., Rustenburg, A. S., Grinaway, P. B., Fass, J., & Chodera, J. D. (2018). Biomolecular simulations under realistic macroscopic salt conditions. The Journal of Physical Chemistry B, 122(21), 5466–5486.
Hess, B., et al. (1997). “AIDJCC4. 3.0”.
Golo, V. L., & Shaĭtan, K. V. (2002). Dynamic attractor for the Berendsen thermostat an the slow dynamics of biomacromolecules. Biofizika, 47(4), 611–617.
Tuble, S. C., Anwar, J., & Gale, J. D. (2004). An approach to developing a force field for molecular simulation of martensitic phase transitions between phases with subtle differences in energy and structure. Journal of the American Chemical Society, 126(1), 396–405.
Páll, S., & Hess, B. (2013). A flexible algorithm for calculating pair interactions on SIMD architectures. Computer Physics Communications, 184(12), 2641–2650.
Kumari, R., & Kumar, R. (2014). C. Open source drug discovery and A. Lynn. Journal of Chemical Information and Modeling, 54(1951), 10–1021.
Liu, K., & Kokubo, H. (2017). Exploring the stability of ligand binding modes to proteins by molecular dynamics simulations: A cross-docking study. Journal of chemical information and modeling, 57(10), 2514–2522.
Manke, I. A., Lowery, D. M., Nguyen, A., & Yaffe, M. B. (2003). BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science, 302(5645), 636–639.
Xiaochun, Y., Chini, C. C. S., He, M., Mer, G., & Chen, J. (2003). The BRCT domain is a phospho-protein binding domain. Science, 302(5645), 639–642.
Barchiesi, G., Roberto, M., Verrico, M., Vici, P., Tomao, S., & Tomao, F. (2021). Emerging role of PARP inhibitors in metastatic triple negative breast cancer. Current scenario and future perspectives. Frontiers in Oncology. https://doi.org/10.3389/fonc.2021.769280
Singh, D. D., Parveen, A., & Yadav, D. K. (2021). Role of PARP in TNBC: Mechanism of inhibition, clinical applications, and resistance. Biomedicines, 9(11), 1512.
Telli, M. L., & Ford, J. M. (2010). PARP inhibitors in breast cancer. Clinical Advances in Hematology & Oncology, 8(9), 629–635.
Kim, D., & Nam, H. J. (2022). PARP inhibitors: Clinical limitations and recent attempts to overcome them. International Journal of Molecular Sciences, 23(15), 8412.
Cortesi, L., Rugo, H. S., & Jackisch, C. (2021). An overview of PARP inhibitors for the treatment of breast cancer. Targeted oncology, 16(3), 255–282.
Lee, H. M., & Moon, A. (2016). Amygdalin regulates apoptosis and adhesion in Hs578T triple-negative breast cancer cells. Biomolecules & therapeutics, 24(1), 62.
Kolesarova, A., Baldovska, S., & Roychoudhury, S. (2021). The multiple actions of amygdalin on cellular processes with an emphasis on female reproduction. Pharmaceuticals, 14(9), 881.
Blaheta, R. A., Nelson, K., Haferkamp, A., & Juengel, E. (2016). Amygdalin, quackery or cure? Phytomedicine, 23(4), 367–376.
Zuhra, K., & Szabo, C. (2022). The two faces of cyanide: An environmental toxin and a potential novel mammalian gasotransmitter. The FEBS Journal, 289(9), 2481–2515.
Tsaur, I., Thomas, A., Monecke, M., Zugelder, M., Rutz, J., Grein, T., Maxeiner, S., Xie, H., Chun, F. K. H., Rothweiler, F., Cinatl, J., Michaelis, M., Haferkamp, A., & Blaheta, R. A. (2022). Amygdalin exerts antitumor activity in taxane-resistant prostate cancer cells. Cancers, 14(13), 3111.
Juengel, E., Thomas, A., Rutz, J., Makarevic, J., Tsaur, I., Nelson, K., Haferkamp, A., & Blaheta, R. A. (2016). Amygdalin inhibits the growth of renal cell carcinoma cells in vitro. International Journal of Molecular Medicine, 37(2), 526–532.
Aamazadeh, F., Ostadrahimi, A., Rahbar Saadat, Y., & Barar, J. (2020). Bitter apricot ethanolic extract induces apoptosis through increasing expression of Bax/Bcl-2 ratio and caspase-3 in PANC-1 pancreatic cancer cells. Molecular Biology Reports, 47(3), 1895–1904.
Liczbiński, P., & Bukowska, B. (2018). Molecular mechanism of amygdalin action in vitro: Review of the latest research. Immunopharmacology and immunotoxicology, 40(3), 212–218.
Duracka, M. I. C. H. A. L., et al. “The impact of Amygdalin on the oxidative profile of rabbit testicular tissue”. Proceedings of the International Conference MendelNet. Vol. 23. 2016.
Hawsawi, Y. M., Shams, A., Theyab, A., Abdali, W. A., Hussien, N. A., Alatwi, H. E., Alzahrani, O. R., Oyouni, A. A., Babalghith, A. O., & Alreshidi, M. (2022). BARD1 mystery: tumor suppressors are cancer susceptibility genes. BMC Cancer, 22(1), 1–23.
Fong, P. C., Boss, D. S., Yap, T. A., Tutt, A., Wu, P., Mergui-Roelvink, M., Mortimer, P., Swaisland, H., Lau, A., O’Connor, M. J., & Ashworth, A. (2009). Inhibition of poly (ADP-ribose) polymerase in tumors from BRCA mutation carriers. New England Journal of Medicine, 361(2), 123–134.
Tung, N., Lin, N. U., Kidd, J., Allen, B. A., Singh, N., Wenstrup, R. J., Hartman, A. R., Winer, E. P., & Garber, J. E. (2016). Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. Journal of Clinical Oncology, 34(13), 1460.
Toss, A., Molinaro, E., Venturelli, M., Domati, F., Marcheselli, L., Piana, S., Barbieri, E., Grandi, G., Piombino, C., Marchi, I., & Tenedini, E. (2020). BRCA detection rate in an Italian cohort of luminal early-onset and triple-negative breast cancer patients without family history: when biology overcomes genealogy. Cancers, 12(5), 1252.
Lin, Shumeng, et al. “Amygdalin Induced Mitochondria-Mediated Apoptosis of Lung Cancer Cells via Regulating NF [Formula: see text] B-1/NF [Formula: see text] B Signaling Cascade in Vitro and in Vivo”. The American journal of Chinese medicine.: 1–26.
Chang, H. K., Shin, M. S., Yang, H. Y., Lee, J. W., Kim, Y. S., Lee, M. H., Kim, J., Kim, K. H., & Kim, C. J. (2006). Amygdalin induces apoptosis through regulation of Bax and Bcl-2 expressions in human DU145 and LNCaP prostate cancer cells. Biological and Pharmaceutical Bulletin, 29(8), 1597–1602.
Moradipoodeh, B., Jamalan, M., Zeinali, M., Fereidoonnezhad, M., & Mohammadzadeh, G. (2019). In vitro and in silico anticancer activity of amygdalin on the SK-BR-3 human breast cancer cell line. Molecular Biology Reports, 46(6), 6361–6370.
Albogami, S., & Alnefaie, A. (2021). Role of Amygdalin in Blocking DNA Replication in Breast Cancer In Vitro. Current Pharmaceutical Biotechnology, 22(12), 1612–1627.
Mosayyebi, B., Mohammadi, L., Kalantary-Charvadeh, A., & Rahmati, M. (2021). Amygdalin decreases adhesion and migration of MDA-MB-231 and MCF-7 breast cancer cell lines. Current Molecular Pharmacology, 14(4), 667–675.
Zielińska, A., Płonka-Czerw, J., Nowak, A., & Kuśmierz, D. (2022). Effect of amygdalin on MCF-7, MDA-MB-231 and T-47D breast cancer cells in the in vitro study. Postępy Higieny i Medycyny Doświadczalnej, 76(1), 132–142.
Al-Khafaji, K., & Tok, T. T. (2021). Amygdalin as multi-target anticancer drug against targets of cell division cycle: Double docking and molecular dynamics simulation. Journal of Biomolecular Structure and Dynamics, 39(6), 1965–1974.
Acknowledgements
The authors sincerely thank the management of Vellore Institute of Technology, Vellore and the Council of Scientific and Industrial Reseach (CSIR), New Delhi, for providing all the necessary amenities required for the development of this computational work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest during the course of this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chatterjee, P., Karn, R., Emerson, I.A. et al. Docking and Molecular Dynamics Simulation Revealed the Potential Inhibitory Activity of Amygdalin in Triple-Negative Breast Cancer Therapeutics Targeting the BRCT Domain of BARD1 Receptor. Mol Biotechnol 66, 718–736 (2024). https://doi.org/10.1007/s12033-023-00680-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-023-00680-8