Abstract
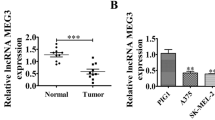
In this study, we aimed to assess the biological functions of HAGLR and its underlying mechanisms in melanoma. HAGLR and ASB11 were knocked down by transfection with the corresponding siRNAs. Meanwhile, miR-4644 was downregulated using the miR-4644 inhibitor treatment. The target interactions among the three molecules were demonstrated using dual-luciferase reporter and RNA immunoprecipitation assays. The levels of HAGLR, miR-4644, and ASB11 in melanoma cells and tissues were assessed using quantitative real‑time PCR and western blotting. The functions and mechanisms underlying HAGLR action in melanoma progression were examined using Cell Counting Kit-8, Transwell, Caspase-3 activity, and xenograft tumor formation assays. HAGLR and ASB11 expression were elevated, whereas that of miR-4644 was downregulated in melanoma cells and tissues. The viability and migration of melanoma cells (A875 and A375) were markedly suppressed by the knockdown of HAGLR and ASB11 but promoted following miR-4644 inhibitor transfection. In contrast, apoptosis showed the opposite trend. In vivo, tumor weight declined considerably with downregulation of HAGLR. Mechanistically, HAGLR sponges miR-4644, increasing the levels of ASB11 and further aggravating melanoma. It latter negatively targets ASB11 in melanoma cells. Hence, the HAGLR-miR-4644-ASB11 axis may be a promising target for melanoma treatment.
Graphical Abstract







Similar content being viewed by others
Data Availability
All data generated and/or analyzed during this research are included in this article.
References
Lodde, G., Zimmer, L., Livingstone, E., Schadendorf, D., & Ugurel, S. (2020). Malignant melanoma. Der Pathologe, 41, 281–292.
Moran, B., Silva, R., Perry, A. S., & Gallagher, W. M. (2018). Epigenetics of malignant melanoma. Seminars in Cancer Biology, 51, 80–88.
Ahmed, B., Qadir, M. I., & Ghafoor, S. (2020). Malignant melanoma: Skin cancer-diagnosis, prevention, and treatment. Critical Reviews in Eukaryotic Gene Expression, 30, 291–297.
Elder, D. E., Bastian, B. C., Cree, I. A., Massi, D., & Scolyer, R. A. (2020). The 2018 World Health Organization classification of cutaneous, mucosal, and uveal melanoma: Detailed analysis of 9 distinct subtypes defined by their evolutionary pathway. Archives of Pathology & Laboratory Medicine, 144, 500–522.
St Laurent, G., Wahlestedt, C., & Kapranov, P. (2015). The landscape of long noncoding RNA classification. Trends in Genetics: TIG, 31, 239–251.
Chi, Y., Wang, D., Wang, J., Yu, W., & Yang, J. (2019). Long non-coding RNA in the pathogenesis of cancers. Cells, 8, 1015.
Tang, L., Zhang, W., Su, B., & Yu, B. (2013). Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. BioMed Research International, 2013, 251098.
Flockhart, R. J., Webster, D. E., Qu, K., Mascarenhas, N., Kovalski, J., Kretz, M., & Khavari, P. A. (2012). BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Research, 22, 1006–1014.
Zhang, Y., Qian, W., Feng, F., Cao, Q., Li, Y., Hou, Y., Zhang, L., & Fan, J. (2019). Upregulated lncRNA CASC2 may inhibit malignant melanoma development through regulating miR-18a-5p/RUNX1. Oncology Research, 27, 371–377.
Wang, X., Wang, Y., Lin, F., Xu, M., & Zhao, X. (2022). Long non-coding RNA LINC00665 promotes melanoma cell growth and migration via regulating the miR-224-5p/VMA21 axis. Experimental Dermatology, 31, 64–73.
Hu, Y. C., Wang, A. M., Lu, J. K., Cen, R., & Liu, L. L. (2017). Long noncoding RNA HOXD-AS1 regulates proliferation of cervical cancer cells by activating Ras/ERK signaling pathway. European Review for Medical and Pharmacological Sciences, 21, 5049–5055.
Li, J., Zhuang, C., Liu, Y., Chen, M., Chen, Y., Chen, Z., He, A., Lin, J., Zhan, Y., Liu, L., Xu, W., Zhao, G., Guo, Y., Wu, H., Cai, Z., & Huang, W. (2016). Synthetic tetracycline-controllable shRNA targeting long non-coding RNA HOXD-AS1 inhibits the progression of bladder cancer. Journal of Experimental & Clinical Cancer Research: CR, 35, 99.
Wang, Y., Zhang, W., Wang, Y., & Wang, S. (2018). HOXD-AS1 promotes cell proliferation, migration and invasion through miR-608/FZD4 axis in ovarian cancer. American Journal of Cancer Research, 8, 170–182.
Zhang, C., Yang, Y., Yi, L., Paizula, X., Xu, W., & Wu, X. (2021). HOXD antisense growth-associated long noncoding RNA promotes triple-negative breast cancer progression by activating Wnt signaling pathway. Journal of Breast Cancer, 24, 315–329.
Li, Y. H., Huang, G. M., Wang, W., & Zang, H. L. (2020). LncRNA HAGLR exacerbates hepatocellular carcinoma through negatively regulating miR-6785-5p. European Review for Medical and Pharmacological Sciences, 24, 9353–9360.
Gu, P., Chen, X., Xie, R., Han, J., Xie, W., Wang, B., Dong, W., Chen, C., Yang, M., Jiang, J., Chen, Z., Huang, J., & Lin, T. (2017). lncRNA HOXD-AS1 regulates proliferation and chemo-resistance of castration-resistant prostate cancer via recruiting WDR5. Molecular Therapy: The Journal of the American Society of Gene Therapy, 25, 1959–1973.
Zhang, H., Bai, M., Zeng, A., Si, L., Yu, N., & Wang, X. (2017). LncRNA HOXD-AS1 promotes melanoma cell proliferation and invasion by suppressing RUNX3 expression. American Journal of Cancer Research, 7, 2526–2535.
Wang, J. Y., Yang, Y., Ma, Y., Wang, F., Xue, A., Zhu, J., Yang, H., Chen, Q., Chen, M., Ye, L., Wu, H., & Zhang, Q. (2020). Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie, 121, 109627.
Cao, D., Wang, Y., Li, D., & Wang, L. (2019). Reconstruction and analysis of the differentially expressed IncRNA-miRNA-mRNA network based on competitive endogenous RNA in hepatocellular carcinoma. Critical Reviews in Eukaryotic Gene Expression, 29, 539–549.
Yan, L., Li, Q., Sun, K., & Jiang, F. (2020). MiR-4644 is upregulated in plasma exosomes of bladder cancer patients and promotes bladder cancer progression by targeting UBIAD1. American Journal of Translational Research, 12, 6277–6289.
Zhao, J., Zhu, X. C., Wu, X. S., Wang, L., Zhu, C. C., Yang, K., Deng, G. Q., Wang, A., Liu, Y., Jia, W. D., & Zhu, L. (2020). Identification of miR-4644 as a suitable endogenous normalizer for circulating miRNA quantification in hepatocellular carcinoma. Journal of Cancer, 11, 7032–7044.
Lei, R., Feng, L., & Hong, D. (2020). ELFN1-AS1 accelerates the proliferation and migration of colorectal cancer via regulation of miR-4644/TRIM44 axis. Cancer Biomarkers: Section A of Disease Markers, 27, 433–443.
Shen, Y., Pan, Y., Xu, L., Chen, L., Liu, L., Chen, H., Chen, Z., & Meng, Z. (2015). Identifying microRNA-mRNA regulatory network in gemcitabine-resistant cells derived from human pancreatic cancer cells. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine, 36, 4525–4534.
Tee, J. M., Sartori da Silva, M. A., Rygiel, A. M., Muncan, V., Bink, R., van den Brink, G. R., van Tijn, P., Zivkovic, D., Kodach, L. L., Guardavaccaro, D., Diks, S. H., & Peppelenbosch, M. P. (2012). asb11 is a regulator of embryonic and adult regenerative myogenesis. Stem Cells and Development, 21, 3091–3103.
Diks, S. H., Bink, R. J., van de Water, S., Joore, J., van Rooijen, C., Verbeek, F. J., den Hertog, J., Peppelenbosch, M. P., & Zivkovic, D. (2006). The novel gene asb11: A regulator of the size of the neural progenitor compartment. The Journal of Cell Biology, 174, 581–592.
Li, J. H., Liu, S., Zhou, H., Qu, L. H., & Yang, J. H. (2014). starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Research, 42, D92-97.
Lewis, B. P., Burge, C. B., & Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120, 15–20.
Melixetian, M., Pelicci, P. G., & Lanfrancone, L. (2022). Regulation of LncRNAs in melanoma and their functional roles in the metastatic process. Cells, 11, 577.
Zheng, L., Chen, J., Zhou, Z., & He, Z. (2017). Knockdown of long non-coding RNA HOXD-AS1 inhibits gastric cancer cell growth via inactivating the JAK2/STAT3 pathway. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine, 39, 1010428317705335.
Qu, Y., Zheng, S., Kang, M., Dong, R., Zhou, H., Zhao, D., & Zhao, J. (2018). Knockdown of long non-coding RNA HOXD-AS1 inhibits the progression of osteosarcoma. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie, 98, 899–906.
Lu, S., Zhou, J., Sun, Y., Li, N., Miao, M., Jiao, B., & Chen, H. (2017). The noncoding RNA HOXD-AS1 is a critical regulator of the metastasis and apoptosis phenotype in human hepatocellular carcinoma. Molecular Cancer, 16, 125.
Paraskevopoulou, M. D., & Hatzigeorgiou, A. G. (2016). Analyzing MiRNA-LncRNA interactions. Methods in Molecular Biology (Clifton, N.J.), 1402, 271–286.
Yang, C., Shen, S., Zheng, X., Ye, K., Sun, Y., Lu, Y., & Ge, H. (2019). Long noncoding RNA HAGLR acts as a microRNA-143-5p sponge to regulate epithelial-mesenchymal transition and metastatic potential in esophageal cancer by regulating LAMP3. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 33, 10490–10504.
Sun, W., Nie, W., Wang, Z., Zhang, H., Li, Y., & Fang, X. (2020). Lnc HAGLR promotes colon cancer progression through sponging miR-185-5p and activating CDK4 and CDK6 in vitro and in vivo. OncoTargets and Therapy, 13, 5913–5925.
Jin, L., Luo, C., Wu, X., Li, M., Wu, S., & Feng, Y. (2021). LncRNA-HAGLR motivates triple negative breast cancer progression by regulation of WNT2 via sponging miR-335-3p. Aging, 13, 19306–19316.
Machida, T., Tomofuji, T., Maruyama, T., Yoneda, T., Ekuni, D., Azuma, T., Miyai, H., Mizuno, H., Kato, H., Tsutsumi, K., Uchida, D., Takaki, A., Okada, H., & Morita, M. (2016). miR-1246 and miR-4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncology Reports, 36, 2375–2381.
Pu, M., Chen, J., Tao, Z., Miao, L., Qi, X., Wang, Y., & Ren, J. (2019). Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cellular and Molecular Life Sciences: CMLS, 76, 441–451.
Guan, Y., Liu, L., Jia, Q., Jin, X., Pang, Y., Meng, F., Zhang, X., & Shen, H. (2020). The role of cell growth-related gene copy number variation in autoimmune thyroid disease. Biological Trace Element Research, 195, 409–416.
Funding
None.
Author information
Authors and Affiliations
Contributions
LJL conceived and designed this study. WHZ formed the methodology. ZL executed the experiments and analysis of data. LJL and WHZ performed the investigations. ZL drafted the paper. This manuscript was reviewed and revised by LJL and WHZ. This work has been reviewed and approved by all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interests.
Consent for Publication
Consent for publication was acquired from the participants.
Informed Consent
All patients signed the written informed consent.
Research Involving Human and/or Animal Participants
The Clinical Ethics Committee of The Third Hospital of Wuhan (Wuhan, China) granted approval to the present study. The processing of clinical tissue specimens was executed in strict compliance with the ethical standards of the Declaration of Helsinki. All patients signed the written informed consent. The execution of the animal experiment strictly observed the ARRIVE guidelines and was authorized by the Animal Ethics Committee of The Third Hospital of Wuhan.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Luo, L., Zhang, W. & Li, Z. LncRNA HAGLR May Aggravate Melanoma Malignancy Via miR-4644/ASB11 Pathway. Mol Biotechnol 65, 1619–1631 (2023). https://doi.org/10.1007/s12033-023-00672-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-023-00672-8