Abstract
Persistence and prevalence of microbial diseases (pandemics, epidemics) is the most alarming threats to the human resulting in huge health and economic losses. Rapid detection and understanding of the disease dynamics by molecular biotechnology tools allow for robust reporting, treatment and control of diseases. As per WHO, the optimal diagnostic approach should be quick, specific, sensitive, without a stringed instrument, and low cost. The drawbacks of traditional detection techniques promote the use of CRISPR-mediated nucleic acid detection methods such as SHERLOCK as detection method. It takes advantage of the unexpected in vitro features of CRISPR-Cas system to develop field-deployable sensitive detection tools. Previously, CRISPR-mediated diagnostic methods have extensively been reviewed particularly for SARS-COV-2 detection, but it fails to provide the insight into advances of this technique. This study is the first attempt to review the advances of SHERLOCK approach as diagnostic tool for viral diseases detection. Variations of SHERLOCK mechanism for improved efficiency are discussed. Particularly integrated SHERLOCK approaches in terms of extraction-free assay and Bluetooth-enabled detection are reviewed to access their feasibility for the development of simpler and cost-effective diagnostic toolkits. Insight in to perks and limitations of diagnostic methods indicates its potential as ultimate diagnostic instrument for disease management.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, the spread of infectious diseases like severe acute respiratory syndrome (SARS), ZIKA, Ebola, Hepatitis, Influenza, Tuberculosis, Human Monkey Pox, Hemorrhagic Fever, Typhoid, etc. has increased, posing a serious threat to public health [1]. The pattern of reoccurrence and resurgence of infectious disease would continue to grow due to increased population, increased number of aged persons, and enhanced inter/intra continental traveling which facilitates the growth, mutation, and spread of novel infectious diseases [2]. In the last 3 decades, the world has witnessed the emergence of more than 30 new infectious diseases. Unfortunately, in most instances, Asia is the epicenter for the emergence and reemergence of these diseases [3]. All these atrocious diseases significantly affected global health, social stability, and the world economy. Authorities in the healthcare system highlighted the importance of screening as many people as possible to detect affected persons and locating their connections as an efficient solution for controlling viral transmission [2].
As per an estimation, until now approximately 39 diagnostic tests have been approved by FDA which mainly rely on laboratory procedures [4]. The current paradigm of clinical disease detection relies on the pathogen culture and detection of specific antibodies, antigens, or nucleic acid of pathogens for which patient samples have to be sent to centralized diagnostic laboratories for analysis [5, 6]. Conventional methods like “Enzyme-Linked Immunosorbent Assay” (ELISA), “Reverse Transcriptase Quantitative PCR” (RT-qPCR), “Next Generation Sequencing” (NGS), and Point of Care (POC) diagnosis are the most common in practice (Table 1) for the microbial identification and nucleic acid testing. However, these methods are time-consuming, rely on laboratory procedures, and require high-cost machines which limit their applicability as on-site diagnostic procedures [5]. It demands the development of robust, efficient, sensitive, accurate, and economical detection methods to enhance the timely disease detection for their intervention.
Alternatively, “Clustered Regularly Interspaced Short Palindromic Repeats” (CRISPR) has emerged as a potent tool for genome editing since its discovery in 1987. The ability of CRISPR and CRISPR-Associated Proteins (Cas Proteins) to correct genetic defects and prevent disease spread enabled scientists to utilize this technology in therapeutics and molecular diagnostics of infectious diseases. These methods have higher diagnostic accuracy and emerged as a promising tool not only for diagnostic purposes but also to identify antibacterial or antiviral-resistant microbes and for the inactivation of infectious pathogens [19]. Mostly the CRISPR-Cas platforms are developed on Cas-9 protein which transformed the world of genetic manipulation thus, speeding the development of new techniques in fundamental studies and clinical practices. CRISPR-Cas9 techniques were able to detect the Zika virus in 2016 and MRSA in 2017 [20]. However, the recognition and cleavage activity is restricted in specific systems because of the requirement of “Protospacer Adjacent Motif” (PAM) or “Protospacer Flanking Sequence” (PFS) [21]. The discovery of Cas12a (formerly Cpf1) and Cas13a (formerly C2c2) proteins revolutionized nucleic acid detection due to their collateral cleavage activity and the lack of PAM and PFS in some orthologs. Particularly Cas13a is ssRNA-specific single component RNA-guided RNA-targeting effector protein that target and cleaves the desired RNA as well as neighboring non-targeted RNA [22].
The particular activity of Cas13a was considered to develop an effective CRISPR platform known as “Specific High-Sensitivity Enzymatic Reporter Unlocking” (SHERLOCK) in 2017 by Prof. Feng Zhang and his team. Later it was used as a diagnostic platform in 2018 for the speedy detection of infectious diseases such as; ZIKA and COVID-19 [17]. In SHERLOCK, nucleic acid pre-amplification was combined with the collateral activity of Cas13 for specific recognition of desired nucleic acid that allows the ultrasensitive, portable, and multiplexed identification of pathogenic nucleic acid from clinical samples [23]. It emerged as promising diagnostic technology, especially in the recent wave of COVID-19. The sensitivity and accuracy of SHERLOCK are advantageous for the development of the cost-effective off-site diagnostic procedure (Table 1) as compared to PCR-based methods. While previously published plethora of literature reviews the CRISPR as diagnostic and therapeutic tool specially for SARS-COV2, the present study reviews the developments of SHERLOCK (CRISPR-mediated technique) and its use as diagnostic tool for viral disease detection. It is first study where the potential and advances of SHERLOCK has comprehensively been reviewed. The objective of this paper is to explain the features, functions, and possibilities of SHERLOCK as prospective testing equipment for infectious viral disease diagnosis to curb the disease outbreaks. Being progressing technique currently SHERLOCK is facing some challenges/shortcomings that are being addressed by integrating other diagnostic methods with this technique. The highlight of the review is the advancements in the SHERLOCK through integrated approaches particularly focusing on extraction-free assays and Bluetooth-enabled detection. This information paves the way for the development of SHERLOCK-based point-of-care devices for viral disease diagnosis.
Mechanism of SHERLOCK-Mediated Approach
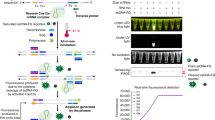
SHERLOCK is the CRISPR-Cas13-based real-time nucleic acid detection method based on the “Recombinase Polymerase Amplification” (RPA) and “Loop-Mediated Isothermal Amplification” (LAMP) amplification of the nucleic acid followed by collateral cleavage of a reported sequence by Cas13a (Fig. 1). Although the original protocol focused on the RPA and LwCas13a, it can be modified for Cas12a, Cas13a, or any other Cas protein variant accordingly. Depending on the choice of reporter molecule, fluorescence detection or lateral flow assay both are compatible with SHERLOCK. Generally, CRISPR-Cas13 system comprises of two components (i) RNA-guided Cas13 RNase with HEPN (higher eukaryotic and prokaryotic nucleotide-binding domain) mediated endonuclease activity, and (ii) pre-CRISPR-RNA (crRNA) that can recognize mature crRNA for targeted activity [24].
Schematic overview of the SHERLOCK-mediated disease diagnostic approach representing the various methods adopted for sample preparation, nucleic acid extraction, and Cas-mediated detection. The figure showed that nucleic acid could be extracted from blood, urine, or nasopharyngeal samples by kit-based extraction, HUDSON method, and magnetic-bead-based extraction. The extracted nucleic acid is then amplified either by LAMP or RPA methods, where amplicons are detected by probe-associated CRISPR-Cas. The results could be obtained on lateral flow strip or through fluorometer
SHERLOCK detection approach consist of three major steps including (i) sample acquisition, (ii) amplification, and (iii) CRISPR-mediated collateral activity and detection. The detection procedure starts from the sample acquisition which could be in the form of blood, urine, and nasal swab or the nucleic acid (either DNA or RNA) extracted from the patient samples. It is followed by the isothermal amplification of the target genome through recombinase polymerase (RPA) or loop-mediated isothermal amplification (LAMP). The target molecules are then subsequently converted to RNA targets by T7 transcription. Binding and collateral cleavage activity of Cas13 is then utilize for the detection. Specificity of this system is ensured by the crRNA-target pairing which is confirmed by designing target-specific Cas13: crRNA complexes according to the target agent to activate and cleave fluorescent RNA sensors. Signal amplification of cleavage activity of LwCas13a protein on RNA reporter (already added in reaction) is done to achieve further sensitivity [17, 25]. The results of collateral activity conferred by detached biotin-fluorescein RNA reporter is then captured. Single-plex colorimetric or lateral flow reactions and single- or multiplex fluorescence-based SHERLOCK reactions are both viable options for detection (Fig. 1). The intensity and existence of fluorescent light indicate the concentration of the target molecule in the clinical samples [25]. Combining Cas13 with Cas12 in the same assay allows for the simultaneous detection of several targets inside the same reaction.
SHERLOCK-Mediated Disease Diagnosis
SHERLOCK has proven as versatile and robust method for the detection of DNA or RNA and has vast applications in rapid disease diagnosis and genotyping. The use of handheld spectrometers or portable electronic fluorescent readers can reduce the instrumentation cost to < 200$ which could be further reduced to 0.61$/test by redesigning the method on paper [25]. In 2017, SHERLOCK v1 was introduced that relies on the Cas13a nuclease originating from Leptotrichia wadei that could detect and cleave RNA molecules unlike the activity of Cas12. Though, SHERLOCK v1 was reliable and able to detect low viral concentrations its applicability was limited by its qualitative nature, dependence on the fluorescent detection instrument for expression readout, and detection of a single target molecule in a single reaction. These limitations were addressed in the SHERLOCK v2 which focused on the (i) combined activity of Cas13 with Csm6 to enhance the sensitivity by 3.5-fold, (ii) 4-channeled multiplexed reaction by Cas enzyme orthologs for the detection of up to four different targets in a single reaction, (iii) quantitative measurement of signals, and (iv) lateral flow readout for instrument-free portable detection. The combination of Cas13 with Csm6 not only improves the sensitivity but also allows the direct detection of most pathogen variants without pre-amplification. These modifications broaden the quantification aspect and the process utility, so it is more visual and sensitive [26]. The validity of SHERLOCK testing for SARS-COV-2 detection on the clinical patient samples from Thailand indicated 100% specificity and sensitivity in the case of fluorescent detection while, lateral flow assay showed 100% specificity with 97% sensitivity [27].
Two-step SHERLOCK testing in combination with LAMP assay was given emergency use authorization by FDA as a portable diagnostic kit for COVID testing thus making it the first-ever FDA-approved CRISPR-Cas diagnostic system. The commercial availability of the kit allows viral detection in one hour with standard laboratory equipment such as a water bath and fluorometer. Testing of the kit on 30 positive and negative nasopharyngeal samples indicated 100% sensitivity and specificity of the test with 6.75 copies/µL of limit-of-detection [28]. However, the cost and the requirement of proper lab setup are the limitations that must be addressed for the economical implementation of such a system in laboratory settings.
Time consumption and higher risks of cross-contamination hinder the applicability of two-step diagnosis. An important variation in this regard enabled the performance of the whole SHERLOCK assay in a single tube which is termed SHERLOCK Testing in One Pot (STOP). It allows for rapid viral detection in femtomolar concentration in 10–15 min and attomolar concentration in less than an hour. It reduces the cross-contamination chances and allows cost-effective offsite disease detection [17]. Recently, a similar type of diagnostic approach is used for detecting SARS-COV 2 (termed STOPCOVID v1). The Nucleic Acid is amplified by LAMP at temperatures 55–70 °C is detected through the specific activity of thermostable Cas12b in the presence of reporter molecule. In STOPCOVID v2, the nucleic acid sample was concentrated by magnetic beads that result in 600 times higher yielding input as compared to the CDC test. Overall, the process can detect 100 copies/sample or 33 copies/mL as compared to 1000 copies/mL detected by the CDC-approved RT-qPCR. Although, this process has the advantage of high sensitivity, unextracted sample processing, and one-fluid handling at a single temperature yet its cost must be reduced. Installation of modular heather will help in streamlining the workflow for a simpler process [29].
Quality assurance of the tested method is challenging due to varying sample collection and preparation methods, incomplete understanding of time course viral dynamics, and lack of standard reference. Integrated approaches based on simpler sample preparation and portable analytical procedures are the future exploratory areas for researchers.
Extraction-Free SHERLOCK Analysis
Replacement of diagnostic gold standard “RT-PCR” with more robust and economical CRISPR-mediated methods would require them to be more scalable, efficient, and sensitive for their easier deployment in low-resource settings. Nucleic acid extraction by commercially available kits is the major hurdle in the field deployment of CRISPR diagnostics due to the multi-step processing, and the requirement of expensive chemicals and trained personnel [30]. Eliminating nucleic acid extraction (Fig. 1) can greatly reduce the time and chemical requirements [5]. The pairing of SHERLOCK with direct sample detection is a must for this technique to excel in any context. Raw nucleic acid extraction through mechanical or chemical forces is suitable but it may contaminate the sample with nucleases or PCR inhibitors which can degrade reporter molecules and cause false-positive signals. Pre-treatment of nucleic acid with RNase inhibitors or HUDSON [inactivation of inhibitors by heat and reducing agents such as Tris (2-carboxyethyl) phosphine hydrochloride (TCEP)] is a viable approach to reduce contamination risk [17]. To develop the field-deployable diagnostic system, HUDSON was integrated with SHERLOCK for the detection of ZIKA and Dengue virus. HUDSDON-treated unextracted saliva or urine samples were directly employed for the RPA while, blood samples need 1:3 dilution with phosphate buffer saline to avoid coagulation during the amplification process [31]. Focusing on this aspect, an extraction-free sensitive SARS-COV-2 detection test known as streamline highlighting of Infections to Navigate Epidemics (SHINE) has been developed. In version 1 of the protocol, patient samples were treated with a mixture of TCEP, EDTA, and murine RNase inhibitors at 40 °C for 5 min followed by incubation at 70 °C for 5 min for sample lysis. The extracted sample was then processed and analyzed by optimized single-step SHERLOCK where visual readouts were monitored through a specially designed smartphone app. The overall process took almost 50 min. Validation of protocol on 50 patients indicates 90% sensitivity and 100% specificity compared with RT-PCR [5]. The process was further simplified in SHINE.v2 by eliminating the need for heating and cold storage of reagents. The modified process took < 90 min and showed 100% specificity and 90.5% of sensitivity as compared to RT-PCR. SHINE.v2 was effectively discriminated between a different variant of the SARS-COV-2. Heat block working at 37 °C and lateral flow sticks were mainly required for the process which enabled its easy use in far reached low-resource areas [32].
Similarly, an RNA-extraction-free CRISPR detection method known as Diagnostics with Coronavirus Enzymatic Reporting (DISCoVER) has been developed for the covid diagnosis. It allows the direct lysis of saliva samples through heating in lysis buffer followed by sample loading in a gravity-driven microfluidic cartridge where samples are amplified by LAMP at 65 °C for 20–30 min. LbuCas13 molecules then identify and cleave the crRNA-specific amplicons at 37 °C and provide fluorescent readouts in 5 min. Unextracted samples are usually at a high risk of degradation due to the presence of RNase in the raw sample as was observed in the present scenario but the treatment of saliva samples with low concentrations of TCEP-EDTA was found to preserve the target RNA. The attomole sensitivity with 100% positive predictive value and 93.7% negative predictive value as seen by automated processes substantially decreases the likelihood of contamination spread. The clinical trial showed 94% sensitivity and 100% specificity of the test [33]. However, the need for specialized equipment is a major limitation of the process in its applicability as a portable diagnostic tool.
Recently, SHERLOCK testing has been optimized with LAMP amplification. Even though, RPA has been in more practice as many CRISPR-based diagnostic methods are developed on it. However, it faces the challenges of cross-contamination due to two-step reactions, weak signals, and long-incubation time. Alternatively, LAMP has a high yield, affordability, and accessibility as it is more specific and speedier [34] but it is limited by false-positive results due to contamination, formation of primer dimers, and sequence similarities [35]. Diseased samples were mixed with proteinase K and heated at 65 °C for 6 min followed by 3 min of treatment at 98 °C. Cooled samples were then processed for the LAMP-based SHERLOCK where amplification was performed at 61 °C for 30 min followed by LwCas13a detection reaction with amplicons at 37 °C. Positive samples were detected by measuring fluorescence for 10 min at 2 min intervals [36].
Integrated Approaches for SHERLOCK Diagnosis
The limitations of the SHERLOCK-mediated diagnosis call for the integration of various methods to enhance the efficiency and efficacy of this technology. Recently, many research groups were able to combine two or more aspects of a process in a single process pipeline to develop efficient methods like SHINE, Sherlock, CARMEN, CREST, etc. which enabled robust disease detection (Fig. 2). Each method has its own set of possibilities and challenges.
Comparative evaluation of SHERLOCK-mediated diagnostic approaches in terms of experimental duration. The figure showed the time required for extraction, amplification, and detection step for various variations of SHERLOCK method. Two-step SHERLOCK took almost 220 min while STOPCovid V2 took almost 60–70 min
In an attempt to develop an economical and self-contained POC diagnostic approach, SHERLOCK was modified to be operated by utilizing minimum instruments (miSHERLOCK). This platform integrated STOP reaction with RNA paper-capture technique for the instrument-free, battery-operated multiplexed detection of SARS-COV-2 and its different variants. The experiment was performed in a specially designed battery-operated heater that could achieve temperatures up to 95 °C. Saliva samples were inactivated at 95 °C and RNA was concentrated onto a column filter which was then automatically plunged into SHERLOCK reaction mixture (consisting of RPA reagents and Cas12a) and processed at 37 °C. Reaction output was monitored visually or through the smartphone app. RNA filtration and concentration through this method not only reduces the chances of cross-contamination but also improves the sensitivity of the assay by 2–20-folds. The cost of miSHERLOCK device is approximately $15 which can further be reduced to $11 by reusing the heater and electronics. The major share of the cost accounts for the commercially obtained reagents and enzymes. Testing of the assay on a small set of saliva samples and the unavailability of test clinical samples were the major limiting factors of the assay [37].
The development of affordable and portable diagnostic facilities is the major goal of the current efforts. Integration of CRISPR with battery-operated, Bluetooth-enabled, field-ready mini thermocyclers allows the deployment of this technology in unconventional environments for viral detection. Based on this aspect, CREST protocol has been developed where the use of mini-PCR mini16 has been suggested as an economical solution for viral target amplification. Whereas for the visualization of results, a battery-powered P51 cardboard fluorescence visualizer has been used instead of lateral flow assay or the fluorometer [38]. Currently, RNA extraction through commercial kits is the limiting factor in the affordable scalability of this technique. An extraction-free protocol precipitation-enhanced analyte retrieval (PEARL) has been combined with CREST to omit the need for commercial RNA extraction kits. PEARL utilize common laboratory reagents such as sodium acetate, TCEP, IGEPAL, polyacrylamide, and HEPES–KOH as lysis solution for nucleic acid or protein extraction. Sample purification was performed by ice-cold precipitation by isopropanol and ethanol. The extracted sample can directly be utilized for detection purposes making the process more streamlined and robust [39].
Routine laboratory diagnostics are sufficient to provide information to patients and healthcare workers to curb the disease’s spread. However, multiplexing of the detection method is essential for robust and inexpensive identification of circulating pathogens [4]. Cas13 approaches are blessed with the advantage of multiplexing which allows the (i) targeting of multiple sites in a single genome, (ii) differentiation between related serotypes and viral species, and (iii) simultaneous detection of multiple mutations within co-circulating SARS-COV-2 variants with high fidelity [24]. Combining the strengths of CRISPR with multiplexing will be ideal for the surveillance of hundreds of samples in one go. For this purpose, the “Combinatorial Arrayed Reactions for Multiplexed Evaluation of Nucleic-acids” (CARMEN) approach was developed where CRISPR-related detecting reagents including Cas13, crRNA, and reporter molecule were combined as fluorescent color-coded nanoliter droplets on the microwell-array system which were then paired with amplified nucleic acid samples in a single-step. Detection reaction monitored by fluorescence microscopy [4].
In general, regardless of the adopted method, there must be some modifications in SHERLOCK or CRISPR-based techniques for their improvement. It includes the (i) development of extraction-free methods with concomitant advances in customized reagents, (ii) improvement in one-pot assays for better control over reaction dynamics, (iii) focus on the portable deployment of the diagnostic methods by improving reagent shelf-life or by employing battery-operated equipment, (iv) identification of novel Cas variants to meet the specific diagnostic needs, (v) per sample reduction in diagnostic cost.
CRISPR Diagnostics and Its Potential Applications
An ideal diagnostic test would have high specificity and low false rates for detecting pathogens, in addition, to be cheap, portable, and capable of differentiating between various types of pathogens. Currently, such test type of diagnostic tolls is very fewer in the market. For the overwhelming infectious as well as noninfectious diseases around the globe, the development of novel instruments that meet the requirements of the WHO’s recommended screening procedures has the potential to profoundly transform epidemiological monitoring and healthcare delivery. Employing a universal “16S rRNA gene V3 RPA” primer set, SHERLOCK can distinguish bacterial strains. SHERLOCK has also been used to uncover antibiotic resistance genes to detect and genotype bacterial or viral infectious disease pathogens. When a critical SNP is known, SHERLOCK can be used to do SNP screening by carefully constructing a crRNA to target the area containing the SNP of interest, which promotes preferential binding of one mutant over another [25].
SHERLOCK has recently been extensively used for the detection of viral diseases such as COVID-19, Dengue, Pneumonia, Hepatitis, Zika, Ebola, and Influenza, etc. (Table 2). In first of its kind study to identify the Zika virus (ZIKV) to determine its genotype, Pardee et al. amplified the viral RNA employing isothermal amplification. The DNA was subsequently cut using CRISPR/Cas9, and this activity was recognized by toehold switch sensors, resulting in a colorimetric display on a test paper. With a single-base precision, this approach can discriminate between the American and African variants of the ZIKA virus [40].
The CRISPR/Cas9 driven isothermal exponential amplification reaction (CAS-EXPAR) technique was recently established by Huang and colleagues. The technique is used to detect the sequence of DNA with attomolar (aM) level of sensitivity & single-base specificity. The target DNA fragment left over after CRISPR/Cas9 cleavage primes CAS-EXPAR, which then cycles through the amplification process to produce a huge number of DNA copies that are identified by utilizing a real-time SYBR Green detectable fluorescent signal. The technique successfully shows that it is sensitive enough to detect Listeria monocytogenes RNA and Epigenetic changes [49]. Another team created two techniques called CRISPR-associated reverse PCR (CARP) and CRISPR-typing PCR (ctPCR) for finding DNA [50]. These tests combine PCR amplification of the target sequence with CRISPR-specific cleavage, and the outcome can subsequently be seen using gel electrophoresis or real-time PCR. Human papillomavirus (HPV) HPV16 and HPV18 L1 gene detection was used to validate ctPCR and CARP. CRISPR/Cas9-based lateral flow assay (CASLFA) was developed more recently by Wang et al. This method combines CRISPR/Cas9 with the lateral flow assay. The CASLFA is used to diagnose Listeria monocytogenes [51] as well as the African swine fever with great specificity and within one hour at a LOD of hundreds of copies of genomic samples.
Another tool that has been modified is the nuclease-dead Cas9 (dCas9) that binds DNA without splitting it. The researchers divided luciferase and joined the two halves to dCas9; these two fragments can heterodimerize to rebuild complete protein and emit bioluminescent signal if targeted to neighboring DNA locations. To diagnose Mycobacterium tuberculosis DNA, the bacterial DNA sequence was pre-amplified, and the dual gRNAs targeting the bacterial DNA were added. If the amplified DNA is the predicted pathogen DNA, bioluminescence will result [49]. A similar technique known as the rolling circle amplification (RCA)-CRISPRsplit-HRP system is employed for quickly detecting MicroRNAs [52]. Pretty recently, Kyeonghye Guk and associates offer a CRISPR-based DNA-FISH approach for the straightforward, quick, and highly sensitive detection employing SYBR Green-I acting as fluorescent probe and dCas9 for precise targeting. Methicillin resistant Staphylococcus aureus (MRSA) can be found using this CRISPR-based DNA-FISH in just 30 min [53].
To increase sensitivity, all of the methods mentioned above need the pre-amplification procedure. The CRISPR-Chip, a label-free DNA-testing device, was created by Reza Hajian et al. to find a target DNA sequence. The biosensor makes use of a field-effect transistor based on graphene and immobilized dCas9-gRNA complex, whose output signal may be detected by a straightforward handheld reader. The CRISPR-Chip can identify mutations in DNA in patients of Duchene muscular dystrophy with a sensitivity of 1.7 femto moles in 15 min. The use of CRISPR-Chip technology to detect nucleic acids electrically on a chip broadens the applicability of CRISPR-Cas9 [54]. In future, exploitation of Cas9 as diagnostic agent in SHERLOCK method will be an interesting avenue to explore.
In another attempt, the improved SHERLOCKv2 utilize the multiplexing approach to enhance the process efficiency. In this technique, four different DNA or RNA sequences can be detected in a single reaction by utilizing a combination of four different Cas13 and Cas12 enzymes. Additionally, the detection sensitivity can be increased by approximately 3.5-fold through the utilization of Csm6, a CRISPR-associated enzyme, in order to boost Cas13 activity. The final step involves the incorporation of a reporter molecule for a lateral flow assay, which enables a visible reading on the test strip. Through the use of lateral flow, SHERLOCKv2 is able to identify mutations in clinical samples as well as single-stranded RNA from the Dengue or Zika virus [26].
Cas12-based monitoring system Cas12a, which is also designated as Cpf1, is a Class V CRISPR-Cas protein. It comprises of one RuvC domain and may identify T-rich PAM sequences for target cleavage as mentioned in Table 3. Because of its ability to degrade dsDNA in the presence of an attached dsDNA substrate, this enzyme also exhibits generalized ssDNase activity. In the DETECTR and HOLMES experiments, Isothermal amplification of the target sequence is performed by RPA or RT-RPA, which subsequently bind the Cas12a-crRNA complex and cause the breakdown of a ssDNA fluorophore-quencher reporter, so producing a fluorescent signal. Both DETECTR and HOLMES are capable of detecting DNA segments with a sensitivity that is attomolar and a specificity that is extremely high [55]. The same approach could be adopted with SHERLOCK to enhance its specificity. Cas12b has an activity that is comparable to that of Cas12a and is utilized in HOLMESv2 and CDetection. CDetection has demonstrated the ability to directly detect HPV16 DNAs in patient blood at concentrations as low as 1 attomolar [56]. There have been improvements to Cas12-based nucleic acid identification. To this day, many amplification techniques such as PCR, RPA, and LAMP and single output forms such as fluorescence detectors, naked-eye views, and subsequently flow assays have been combined into the procedures, which has allowed them to become sensitive, accurate, portable, and simple to employ [49].
One of the tiniest Cas proteins, Cas14, contains 24 distinct variants grouped into three distinct families. Proteins belonging to the Cas14 family are able to breakdown the targeted DNA sequence without the need for needing a PAM in order to be activated. Furthermore, target identification by Cas 14 triggers the breakdown of ssDNA molecule that is not specific. Cas14a has been incorporated into the DETECTR platform so that it can create a brand-new system for the detection of ssDNA called Cas14a-DETECTR. It makes possible high-fidelity DNA SNP analysis (detection of single-nucleotide polymorphisms) without being constrained by the PAM. Because of this inspirational study, a number of scientists have proposed analyzing DNA as part of their work. Cas14a-DETECTR is able to detect pathogens, but there has been no documented case of the diagnosis of a pathogen [58]. In future, research on the suitability of Cas14 as diagnostic marker with SHERLOCK method will be an interesting avenue to explore.
Perks and Pitfalls of CRISPR/Cas-Mediated Pathogen Detection Systems
Using CRISPR/Cas-based methods, microbes like SARS-CoV-2, Zika, Ebola, HPV, and Mycobacterium tuberculosis can be detected with greater precision, speed, cost-effectiveness, and ease. CRIPSR-mediated system also has the potential to be used as identification, imaging, and therapeutic tool in addition to its use as disease diagnostic approach (Fig. 3). The procedures are undergoing additional enhancements and modifications in order to render them more adaptable [49].
Potential applications of the CRISPR diagnostics for identification, detection, and therapeutics. The figure showed that SHERLOCK is used for species identification and single-nucleotide polymorphism analysis. Nucleic acid detection could be adopted for mutation analysis or antibiotic resistance gene detection. As diagnostic tool, SHERLOCK is used to detect bacterial, viral, and infectious diseases. CRISPR-mediated nucleic acid imaging is useful for disease genotyping. Nucleic acid editing and gene expression regulation by SHERLOCK are the potential areas for its as therapeutic agent
The ease of use of the CRISPR/Cas-based detection system makes it possible to rapidly develop diagnostic methods in the event of an unexpected outbreak of infectious diseases. This is an extremely important feature. For instance, integrated later flow assay (CRISPR/Cas12a-LFD) and naked-eye detection (CRISPR/Cas-based colorimetric system, fluorescence-based POC system) was developed for rapid Cas12a-mediated on-site detection assays of African swine virus (ASFV), an emerging virus for the international swine industry [41]. These screening tools can identify as few as two copies of RNA and readout on a later flow ribbon in 60 min without the need for complicated instrumentation or visualization by green fluorescent with the naked-eye under 485 nm light. In addition, these diagnostic methods can detect as few as two copies of SARS-CoV-2 RNA. While back, Joung and peers [29] proposed an integrated assay for detecting SARS-CoV-2 called STOPCovid (SHERLOCK Testing in One Pot). This test can return result on the strip in 70 min, with an LOD of 100 copies of viral RNA In saliva or nasopharyngeal swabs. These CRISPR/Cas-based SARS-CoV-2 tests, in contrast to the RT-qPCR-based diagnostic methods, have the advantages of high speed, the absence of a requirement for expensive instrumentation and the assistance of trained technician, and will be a significant help in the COVID-19 epidemic control.
SHERLOCK uses the Cas13 enzyme, which does not require tight sequence partialities at the target location, whereas Cas12 requires a PAM to cleave. As a result, SHERLOCK can tolerate a longer target range as compared to DETECTR [59].
Despite its advantages over existing detection systems, SHERLOCK has some drawbacks that may make it unsuitable in some situations. Currently, SHERLOCK entails the manufacture and testing of reaction components, some of which necessitate knowledge of protein purification as well as RNA biology. Furthermore, pre-designed assays for SHERLOCK, such as reaction mixes and DNA/RNA oligonucleotides, are not presently commercially available. Existing routine detection techniques, such as cancer assays, may be better for applications that do not require the speed or portability of SHERLOCK [17].
The multi-step nucleic acid amplification procedure, which may influence precise target quantification, is another possible restriction of SHERLOCK. Although Kellner et al. recently showed quantitative nucleic acid detection using SHERLOCK, absolute digital quantification, such as in digital droplet PCR, is presently not attainable, and minor differences in target quantity may not be identified. As a result, SHERLOCK may be less useful for determining accurate gene expression profiles [17].
Opportunities for SHERLOCK-Mediated Pathogen Detection Systems
The SHERLOCK reaction mixture can be lyophilized and utilized after long periods of storage without affecting the test's sensitivity and specificity. SHERLOCK can easily identify between viruses like the dengue virus and the Zika virus [25]. SHERLOCKv2 can identify two cancer mutations in simulated cell-free DNA (cfDNA) specimens with single-base mismatch sensitivity under low allelic fraction [60]. In parallel to in vitro RNA target identification, catalytically active LwaCas13 preserves its RNA-binding activity, allowing for live cell RNA tracking using a fluorescent probe. This provides a different way of recognizing and visualizing RNA. SHERLOCK’s single-nucleotide specificity has been used to offer a genotyping profile of cancer victims by exposing cancer-associated mutations in circulating cell-free DNA, even at low attomolar concentrations of 0.1% allelic fraction in serum samples or urine samples. Cas13’s specificity can be enhanced in such circumstances by inserting a synthetic mismatch’ into the crRNA [59].
Bio-SCAN (biotin-coupled-specific CRISPR-based assay for nucleic acid detection) was created as an efficient pathogen detection tool that doesn’t require any special equipment or technical knowledge. From specimen collection to result, Bio-SCAN recognizes the SARS-CoV-2 nucleic acid in less than 1 h. In the first phase, the target sequence is isothermally amplified in 15 min using recombinase polymerase amplification (RPA) then precisely identified on commercially available lateral flow strips using biotin-labeled nuclease-dead SpCas9 (dCas9). Furthermore, Bio-SCAN was able to discriminate between the SARS-CoV-2 variants using variant-specific guide RNAs in the detection reaction. The findings also revealed that the Bio-SCAN assay reagents and chemicals have a long shelf-life and can be constructed locally in non-laboratory and low-resource environments. The Bio-SCAN system can also be used with the nucleic acid rapid extraction methodology. Our findings indicate Bio-potential SCAN’s as a potential point-of-care diagnostic technology for cost-effective mass screening for SARS-CoV-2 [61].
Cas13’s specificity can be improved by inserting a synthetic mismatch’ into the crRNA. The assay’s speed is one of SHERLOCK’s most appealing features. RPA is usually done for 5–10 min as a first reaction, after which a portion of the solution is passed to the Cas13 detection reaction, which can detect the target in 5 min. The inexpensive cost of its components is another perk of the SHERLOCK system over other detection methods (such as TaqMan qPCR). A typical single-plex reaction costs around $0.60.
The point-of-care testing (PoC) may get benefit from nanotechnology in the near future. In addition to lateral flow tests, several additional diagnostic procedures like chest computed tomography, other new variants of Cas proteins, and paper microfluidic device can be integrated with smartphones and artificial intelligence (AI) for point-of-care testing. These advancements will completely change the way diseases are diagnosed by providing a POC testing method that is rapid, accurate, low cost, and user-friendly. These technologies need to be developed further so that they can be quickly put into action in the event of unanticipated medical emergency.
Conclusion
SHERLOCK is recently developed CRISPR-mediated diagnostic technique with high sensitivity and specificity. It allows for the single-molecule detection of DNA and RNA target sequences in as low as 1-µL sample volumes without the need of specialized instruments which makes it suitable for on-site disease diagnostics. SHERLOCK takes advantage of both Cas13 and Cas12 enzyme specificity. Integration of SHERLOCK with extraction-free assays, multiplexing with Cas variants, and Bluetooth-enabled detection had ushered a new era in molecular diagnostics by offering portable, extremely sensitive diagnostic platforms capable of quickly diagnosing emerging infectious & non-infectious viral diseases. Albeit still in its infancy, SHERLOCK assay has proved to be a game-changer which have the potential to revolutionize our capability of detecting infectious disease and pathogens with ultra-sensitive tests which do not involve extensive complicated processing, allowing for mass screening and better control of viral outbreaks as well as widespread distribution of field-deployable diagnostics toolkits at an affordable cost.
Abbreviations
- SARS:
-
Severe acute respiratory syndrome
- MRSA:
-
Methicillin resistant Staphylococcus aureus
- FDA:
-
Food and Drug Authority
- CDC:
-
Center of Disease Control
- ELISA:
-
Enzyme-Linked Immunosorbent Assay
- RT-qPCR:
-
Reverse Transcriptase Quantitative PCR
- NGS:
-
Next Generation Sequencing
- POC:
-
Point of Care
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- Cas:
-
CRISPR-associated proteins
- PAM:
-
Protospacer Adjacent Motif
- PFS:
-
Protospacer Flanking Sequence
- HEPN:
-
Higher eukaryotic and prokaryotic nucleotide-binding domain
- RPA:
-
Recombinase polymerase amplification
- LAMP:
-
Loop-mediated isothermal amplification
- SHERLOCK:
-
Specific High-Sensitivity Enzymatic Reporter Unlocking
- STOP:
-
SHERLOCK Testing in One Pot
- TCEP:
-
Tris (2-carboxyethyl) phosphine hydrochloride
- SHINE:
-
Streamline Highlighting of Infections to Navigate Epidemics
- DISCOVER:
-
Diagnostics with Coronavirus Enzymatic Reporting
- CARMEN:
-
Combinatorial Arrayed Reactions for Multiplexed Evaluation of Nucleic-acids
- PEARL:
-
Precipitation-enhanced analyte retrieval
- CAS-EXPAR:
-
CRISPR/Cas9 driven isothermal exponential amplification reaction
- CARP:
-
CRISPR-associated reverse PCR
- ctPCR:
-
CRISPR-typing PCR
- CASLFA:
-
CRISPR/Cas9-based lateral flow assay
- RCA:
-
Rolling circle amplification
- cfDNA:
-
Cell-free DNA
- Bio-SCAN:
-
Biotin-coupled-specific CRISPR-based assay for nucleic acid detection
References
Yuan, K., Huang, X.-L., Yan, W., Zhang, Y.-X., Gong, Y.-M., Su, S.-Z., Huang, Y.-T., Zhong, Y., Wang, Y.-J., Yuan, Z., Tian, S.-S., Zheng, Y.-B., Fan, T.-T., Zhang, Y.-J., Meng, S.-Q., Sun, Y.-K., Lin, X., Zhang, T.-M., Ran, M.-S., … Lu, L. (2022). A systematic review and meta-analysis on the prevalence of stigma in infectious diseases, including COVID-19: A call to action. Molecular Psychiatry, 27, 19–33.
Giri, B., Pandey, S., Shrestha, R., Pokharel, K., Ligler, F. S., & Neupane, B. B. (2021). Review of analytical performance of COVID-19 detection methods. Analytical and Bioanalytical Chemistry, 413, 35–48.
World Health Organization, Regional Office for South-East Asia. (2014). A brief guide to emerging infectious diseases and zoonoses. New Delhi: WHO Regional Office for South-East Asia.
Ackerman, C. M., Myhrvold, C., Thakku, S. G., Freije, C. A., Metsky, H. C., Yang, D. K., Ye, S. H., Boehm, C. K., Kosoko-Thoroddsen, T.-S.F., Kehe, J., Nguyen, T. G., Carter, A., Kulesa, A., Barnes, J. R., Dugan, V. G., Hung, D. T., Blainey, P. C., & Sabeti, P. C. (2020). Massively multiplexed nucleic acid detection with Cas13. Nature, 582, 277–282.
Arizti-Sanz, J., Freije, C. A., Stanton, A. C., Petros, B. A., Boehm, C. K., Siddiqui, S., Shaw, B. M., Adams, G., Kosoko-Thoroddsen, T.-S.F., Kemball, M. E., Uwanibe, J. N., Ajogbasile, F. V., Eromon, P. E., Gross, R., Wronka, L., Caviness, K., Hensley, L. E., Bergman, N. H., MacInnis, B. L., … Myhrvold, C. (2020). Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nature Communications, 11, 5921.
Wang, C., Liu, M., Wang, Z., Li, S., Deng, Y., & He, N. (2021). Point-of-care diagnostics for infectious diseases: From methods to devices. Nano Today, 37, 101092.
D’Cruz, R. J., Currier, A. W., & Sampson, V. B. (2020). Laboratory testing methods for novel severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2). Frontiers in Cell and Developmental Biology, 8, 468.
Green, K., Winter, A., Dickinson, R., Graziadio, S., Wolff, R., Mallett, S., Allen, A. J., & Park, E. (2020). What tests could potentially be used for the screening, diagnosis and monitoring of COVID-19 and what are their advantages and disadvantages. CEBM, 13. https://doi.org/10.1016/j.ejrad.2020.109009.
Krüttgen, A., Cornelissen, C. G., Dreher, M., Hornef, M., Imöhl, M., & Kleines, M. (2020). Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. Journal of Clinical Virology, 128, 104394.
Simner, P. J., Miller, S., & Carroll, K. C. (2018). Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clinical Infectious Diseases, 66, 778–788.
Menon, S., Mathew, M. R., Sam, S., Keerthi, K., & Kumar, K. G. (2020). Recent advances and challenges in electrochemical biosensors for emerging and re-emerging infectious diseases. Journal of Electroanalytical Chemistry, 878, 114596.
Jeddi, I., & Saiz, L. (2017). Three-dimensional modeling of single stranded DNA hairpins for aptamer-based biosensors. Science and Reports, 7, 1178.
Low, S. S., Loh, H.-S., Boey, J. S., Khiew, P. S., Chiu, W. S., & Tan, M. T. T. (2017). Sensitivity enhancement of graphene/zinc oxide nanocomposite-based electrochemical impedance genosensor for single stranded RNA detection. Biosensors and Bioelectronics, 94, 365–373.
Reddy, Y. V. M., Shin, J. H., Palakollu, V. N., Sravani, B., Choi, C.-H., Park, K., Kim, S.-K., Madhavi, G., Park, J. P., & Shetti, N. P. (2022). Strategies, advances, and challenges associated with the use of graphene-based nanocomposites for electrochemical biosensors. Advances in Colloid and Interface Science, 304, 102664.
Brandsma, E., Verhagen, H. J. M. P., van de Laar, T. J. W., Claas, E. C. J., Cornelissen, M., & van den Akker, E. (2020). Rapid, sensitive and specific SARS coronavirus-2 detection: A multi-center comparison between standard qRT-PCR and CRISPR based DETECTR. medRxiv. 2020.2007.2027.20147249.
Broughton, J. P., Deng, X., Yu, G., Fasching, C. L., Singh, J., Streithorst, J., Granados, A., Sotomayor-Gonzalez, A., Zorn, K., Gopez, A., Hsu, E., Gu, W., Miller, S., Pan, C.-Y., Guevara, H., Wadford, D. A., Chen, J. S., & Chiu, C. Y. (2020). Rapid detection of 2019 novel coronavirus SARS-CoV-2 using a CRISPR-based DETECTR lateral flow assay. medRxiv: The Preprint Server for Health Sciences, 2020.2003.2006.20032334.
Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O., & Zhang, F. (2019). SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nature Protocols, 14, 2986–3012.
Selvam, K., Najib, M. A., Khalid, M. F., Mohamad, S., Palaz, F., Ozsoz, M., & Aziah, I. (2021). RT-LAMP CRISPR-Cas12/13-based SARS-CoV-2 detection methods. Diagnostics, 11, 1646.
Safari, F., Afarid, M., Rastegari, B., Borhani-Haghighi, A., Barekati-Mowahed, M., & Behzad-Behbahani, A. (2021). CRISPR systems: Novel approaches for detection and combating COVID-19. Virus Research, 294, 198282.
Ahmad, S., Wei, X., Sheng, Z., Hu, P., & Tang, S. (2020). CRISPR/Cas9 for development of disease resistance in plants: Recent progress, limitations and future prospects. Briefings in Functional Genomics, 19, 26–39.
Kaminski, M. M., Abudayyeh, O. O., Gootenberg, J. S., Zhang, F., & Collins, J. J. (2021). CRISPR-based diagnostics. Nature BioMedical Engineering, 5, 643–656.
Jolany vangah, S., Katalani, C., Boone, H. A., Hajizade, A., Sijercic, A., & Ahmadian, G. (2020). CRISPR-based diagnosis of infectious and noninfectious diseases. Biological Procedures Online, 22, 22.
Mustafa Mujahed, I., Makhawi Abdelrafie, M., & Kraft Colleen, S. (2021). SHERLOCK and DETECTR: CRISPR-Cas systems as potential rapid diagnostic tools for emerging infectious diseases. Journal of Clinical Microbiology, 59, e00745-00720.
Aquino-Jarquin, G. (2021). Recent progress on rapid SARS-CoV-2/COVID-19 detection by CRISPR-Cas13-based platforms. Drug Discovery Today, 26, 2025–2035.
Gootenberg Jonathan, S., Abudayyeh Omar, O., Lee Jeong, W., Essletzbichler, P., Dy Aaron, J., Joung, J., Verdine, V., Donghia, N., Daringer Nichole, M., Freije Catherine, A., Myhrvold, C., Bhattacharyya Roby, P., Livny, J., Regev, A., Koonin Eugene, V., Hung Deborah, T., Sabeti Pardis, C., Collins James, J., & Zhang, F. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science, 356, 438–442.
Gootenberg Jonathan, S., Abudayyeh Omar, O., Kellner Max, J., Joung, J., Collins James, J., & Zhang, F. (2018). Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science, 360, 439–444.
Patchsung, M., Jantarug, K., Pattama, A., Aphicho, K., Suraritdechachai, S., Meesawat, P., Sappakhaw, K., Leelahakorn, N., Ruenkam, T., Wongsatit, T., Athipanyasilp, N., Eiamthong, B., Lakkanasirorat, B., Phoodokmai, T., Niljianskul, N., Pakotiprapha, D., Chanarat, S., Homchan, A., Tinikul, R., … Uttamapinant, C. (2020). Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nature Biomedical Engineering, 4, 1140–1149.
Sherlock-Biosciences. (2020). Instructions for use for Sherlock™ CRISPR SARS-CoV-2 kit. http://www.sherlock.bio/.
Joung, J., Ladha, A., Saito, M., Kim, N.-G., Woolley, A. E., Segel, M., Barretto, R. P. J., Ranu, A., Macrae, R. K., Faure, G., Ioannidi, E. I., Krajeski, R. N., Bruneau, R., Huang, M.-L.W., Yu, X. G., Li, J. Z., Walker, B. D., Hung, D. T., Greninger, A. L., … Zhang, F. (2020). Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. New England Journal of Medicine, 383, 1492–1494.
Ganbaatar, U., & Liu, C. (2021). CRISPR-based COVID-19 testing: Toward next-generation point-of-care diagnostics. Frontiers in Cellular and Infection Microbiology, 11, 663949.
Myhrvold, C., Freije Catherine, A., Gootenberg Jonathan, S., Abudayyeh Omar, O., Metsky Hayden, C., Durbin Ann, F., Kellner Max, J., Tan Amanda, L., Paul Lauren, M., Parham Leda, A., Garcia Kimberly, F., Barnes Kayla, G., Chak, B., Mondini, A., Nogueira Mauricio, L., Isern, S., Michael Scott, F., Lorenzana, I., Yozwiak Nathan, L., … Sabeti Pardis, C. (2018). Field-deployable viral diagnostics using CRISPR-Cas13. Science, 360, 444–448.
Arizti-Sanz, J., Bradley, A. D., Zhang, Y. B., Boehm, C. K., Freije, C. A., Grunberg, M. E., Kosoko-Thoroddsen, T.-S. F., Welch, N. L., Pillai, P. P., Mantena, S., Kim, G., Uwanibe, J. N., John, O. G., Eromon, P. E., Kocher, G., Gross, R., Lee, J. S., Hensley, L. E., MacInnis, B. L., … Myhrvold, C. (2022). Simplified Cas13-based assays for the fast identification of SARS-CoV-2 and its variants. Nature Biomedical Engineering, 6, 932–943.
Agrawal, S., Fanton, A., Chandrasekaran, S. S., Charrez, B., Escajeda, A. M., Son, S., McIntosh, R., Bhuiya, A., de León Derby, M. D., Switz, N. A., Armstrong, M., Harris, A. R., Prywes, N., Lukarska, M., Biering, S. B., Smock, D. C. J., Mok, A., Knott, G. J., Dang, Q., … Hsu, P. D. (2021). Rapid, point-of-care molecular diagnostics with Cas13. medRxiv, 2020.2012.2014.20247874.
Wang, R., Qian, C., Pang, Y., Li, M., Yang, Y., Ma, H., Zhao, M., Qian, F., Yu, H., Liu, Z., Ni, T., Zheng, Y., & Wang, Y. (2021). opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosensors & Bioelectronics, 172, 112766.
Warmt, C., Yaslanmaz, C., & Henkel, J. (2022). Investigation and validation of labelling loop mediated isothermal amplification (LAMP) products with different nucleotide modifications for various downstream analysis. Science and Reports, 12, 7137.
Manning, B. J., Khan, W. A., Peña, J. M., Fiore, E. S., Boisvert, H., Tudino, M. C., Barney, R. E., Wilson, M. K., Singh, S., Mowatt, J. A., Thompson, H. J., Tsongalis, G. J., & Blake, W. J. (2022). High-throughput CRISPR–Cas13 SARS-CoV-2 test. Clinical Chemistry, 68, 172–180.
de Puig, H., Lee Rose, A., Najjar, D., Tan, X., Soenksen Luis, R., Angenent-Mari Nicolaas, M., Donghia Nina, M., Weckman Nicole, E., Ory, A., Ng Carlos, F., Nguyen Peter, Q., Mao Angelo, S., Ferrante Thomas, C., Lansberry, G., Sallum, H., Niemi, J., & Collins James, J. (2021). Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Science Advances, 7, eabh2944.
Rauch Jennifer, N., Valois, E., Solley Sabrina, C., Braig, F., Lach Ryan, S., Audouard, M., Ponce-Rojas Jose, C., Costello Michael, S., Baxter Naomi, J., Kosik Kenneth, S., Arias, C., Acosta-Alvear, D., Wilson Maxwell, Z., & Hayden, R. (2021). A scalable, easy-to-deploy protocol for Cas13-based detection of SARS-CoV-2 genetic material. Journal of Clinical Microbiology, 59, e02402-02420.
Ponce-Rojas Jose, C., Costello Michael, S., Proctor Duncan, A., Kosik Kenneth, S., Wilson Maxwell, Z., Arias, C., Acosta-Alvear, D., & Hayden, R. (2021). A fast and accessible method for the isolation of RNA, DNA, and protein to facilitate the detection of SARS-CoV-2. Journal of Clinical Microbiology, 59, e02403-02420.
Pardee, K., Green, A. A., Takahashi, M. K., Braff, D., Lambert, G., Lee, J. W., Ferrante, T., Ma, D., Donghia, N., Fan, M., Daringer, N. M., Bosch, I., Dudley, D. M., O’Connor, D. H., Gehrke, L., & Collins, J. J. (2016). Rapid, low-cost detection of Zika Virus using programmable biomolecular components. Cell, 165, 1255–1266.
Joung, J., Ladha, A., Saito, M., Segel, M., Bruneau, R., Huang, M.-l. W., Kim, N.-G., Yu, X., Li, J., Walker, B. D., Greninger, A. L., Jerome, K. R., Gootenberg, J. S., Abudayyeh, O. O., & Zhang, F. (2020). Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv. 2020.2005.2004.20091231.
Casati, B., Verdi, J. P., Hempelmann, A., Kittel, M., Klaebisch, A. G., Meister, B., Welker, S., Asthana, S., Di Giorgio, S., Boskovic, P., Man, K. H., Schopp, M., Ginno, P. A., Radlwimmer, B., Stebbins, C. E., Miethke, T., Papavasiliou, F. N., & Pecori, R. (2022). Rapid, adaptable and sensitive Cas13-based COVID-19 diagnostics using ADESSO. Nature Communications, 13, 3308.
Wang, Y., Li, J., Li, S., Zhu, X., Wang, X., Huang, J., Yang, X., & Tai, J. (2021). LAMP-CRISPR-Cas12-based diagnostic platform for detection of Mycobacterium tuberculosis complex using real-time fluorescence or lateral flow test. Microchimica Acta, 188, 347.
Zhan, Y., Gao, X., Li, S., Si, Y., Li, Y., Han, X., Sun, W., Li, Z., & Ye, F. (2022). Development and evaluation of rapid and accurate CRISPR/Cas13-based RNA diagnostics for Pneumocystis jirovecii pneumonia. Frontiers in Cellular and Infection Microbiology, 12, 904485.
Ding, R., Shen, Y., Yuan, M., Zheng, X., Chen, S., & Duan, G. (2022). Rapid and facile detection of HBV with CRISPR/Cas13a. New Journal of Chemistry, 46, 19997–20004.
Barnes, K. G., Lachenauer, A. E., Nitido, A., Siddiqui, S., Gross, R., Beitzel, B., Siddle, K. J., Freije, C. A., Dighero-Kemp, B., Mehta, S. B., Carter, A., Uwanibe, J., Ajogbasile, F., Olumade, T., Odia, I., Sandi, J. D., Momoh, M., Metsky, H. C., Boehm, C. K., … Sabeti, P. C. (2020). Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real-time. Nature Communications, 11, 4131.
Liu, Y., Xu, H., Liu, C., Peng, L., Khan, H., Cui, L., Huang, R., Wu, C., Shen, S., Wang, S., Liang, W., Li, Z., Xu, B., & He, N. (2019). CRISPR-Cas13a nanomachine based simple technology for avian influenza A (H7N9) virus on-site detection. Journal of Biomedical Nanotechnology, 15, 790–798.
Yuan, C., Tian, T., Sun, J., Hu, M., Wang, X., Xiong, E., Cheng, M., Bao, Y., Lin, W., Jiang, J., Yang, C., Chen, Q., Zhang, H., Wang, H., Wang, X., Deng, X., Liao, X., Liu, Y., Wang, Z., … Zhou, X. (2020). Universal and naked-eye gene detection platform based on the clustered regularly interspaced short palindromic repeats/Cas12a/13a system. Analytical Chemistry, 92, 4029–4037.
Wang, X., Shang, X., & Huang, X. (2020). Next-generation pathogen diagnosis with CRISPR/Cas-based detection methods. Emerging Microbes & Infections, 9, 1682–1691.
Zhang, B., Xia, Q., Wang, Q., Xia, X., & Wang, J. (2018). Detecting and typing target DNA with a novel CRISPR-typing PCR (ctPCR) technique. Analytical Biochemistry, 561–562, 37–46.
Wang, X., Xiong, E., Tian, T., Cheng, M., Lin, W., Wang, H., Zhang, G., Sun, J., & Zhou, X. (2020). Clustered regularly interspaced short palindromic repeats/Cas9-mediated lateral flow nucleic acid assay. ACS Nano, 14, 2497–2508.
Qiu, X.-Y., Zhu, L.-Y., Zhu, C.-S., Ma, J.-X., Hou, T., Wu, X.-M., Xie, S.-S., Min, L., Tan, D.-A., Zhang, D.-Y., & Zhu, L. (2018). Highly effective and low-cost microRNA detection with CRISPR-Cas9. ACS Synthetic Biology, 7, 807–813.
Guk, K., Keem, J. O., Hwang, S. G., Kim, H., Kang, T., Lim, E.-K., & Jung, J. (2017). A facile, rapid and sensitive detection of MRSA using a CRISPR-mediated DNA FISH method, antibody-like dCas9/sgRNA complex. Biosensors & Bioelectronics, 95, 67–71.
Hajian, R., Balderston, S., Tran, T., deBoer, T., Etienne, J., Sandhu, M., Wauford, N. A., Chung, J.-Y., Nokes, J., Athaiya, M., Paredes, J., Peytavi, R., Goldsmith, B., Murthy, N., Conboy, I. M., & Aran, K. (2019). Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nature Biomedical Engineering, 3, 427–437.
Chen, J. S., Ma, E., Harrington, L. B., Da Costa, M., Tian, X., Palefsky, J. M., & Doudna, J. A. (2018). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science, 360, 436–439.
Teng, F., Guo, L., Cui, T., Wang, X.-G., Xu, K., Gao, Q., Zhou, Q., & Li, W. (2019). CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biology, 20, 132.
Feng, W., Newbigging, A. M., Tao, J., Cao, Y., Peng, H., Le, C., Wu, J., Pang, B., Li, J., Tyrrell, D. L., Zhang, H., & Le, X. C. (2021). CRISPR technology incorporating amplification strategies: Molecular assays for nucleic acids, proteins, and small molecules. Chemical Science, 12, 4683–4698.
Harrington, L. B., Burstein, D., Chen, J. S., Paez-Espino, D., Ma, E., Witte, I. P., Cofsky, J. C., Kyrpides, N. C., Banfield, J. F., & Doudna, J. A. (2018). Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science, 362, 839–842.
Huang, C.-H., Lee, K.-C., & Doudna, J. A. (2018). Applications of CRISPR-Cas enzymes in cancer therapeutics and detection. Trends in Cancer, 4, 499–512.
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., Verdine, V., Donghia, N., Daringer, N. M., Freije, C. A., Myhrvold, C., Bhattacharyya, R. P., Livny, J., Regev, A., Koonin, E. V., Hung, D. T., Sabeti, P. C., Collins, J. J., & Zhang, F. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science, 356, 438–442.
Ali, Z., Sánchez, E., Tehseen, M., Mahas, A., Marsic, T., Aman, R., Sivakrishna Rao, G., Alhamlan, F. S., Alsanea, M. S., Al-Qahtani, A. A., Hamdan, S., & Mahfouz, M. (2022). Bio-SCAN: A CRISPR/dCas9-based lateral flow assay for rapid, specific, and sensitive detection of SARS-CoV-2. ACS Synthetic Biology, 11, 406–419.
Funding
This study was supported by Ministry of Science and Technology, Pakistan (Grant No. DDWP-2021) to PCSIR and University of Agriculture Faisalabad, Pakistan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zahra, A., Shahid, A., Shamim, A. et al. The SHERLOCK Platform: An Insight into Advances in Viral Disease Diagnosis. Mol Biotechnol 65, 699–714 (2023). https://doi.org/10.1007/s12033-022-00625-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00625-7