Abstract
Acute myeloid leukemia (AML) is a fatal heterogeneous hematologic malignancy. There is an urgent need to identify potential biomarkers to better classify sufferers with bad outcomes that might need more advanced treatment. The objective of this study was to investigate prognostic indicators that predict the outcome of sufferers with AML. The datasets of AML sufferers including mRNA sequencing data and clinical information were acquired from GEO datasets (GSE38865) and TCGA datasets. Kaplan–Meier curves and Cox regression analysis to screen genes correlated to survival. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses biological process analysis were utilized in verifying the function of various genes. Sufferers with elevated MCM5 level exhibited a worse prognosis, according to the survival analysis. It was indicated through multivariate and univariate analysis that MCM5 level was an independent adverse prognostic element for over survival in AML sufferers based on GEO and TCGA datasets. Meanwhile, MCM5 level in AML samples was higher than in normal samples. Additionally, it was indicated through PPI network and functional enrichment analyses that through accelerating cell cycle and DNA replication, MCM5 promoted AML progression. In conclusions, MCM5 level was an independent poor prognostic element in AML sufferers based on GEO and TCGA datasets. This is the first time that MCM5 is reported to be a biomarker of poor prognosis in AML.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
In adults, the most common acute leukemia is acute myeloid leukemia (AML) [1]. AML is a cell clonal malignant proliferative disease of myeloid primordial cells in the hematopoietic system. AML is a highly heterogeneous group of diseases that can be derived from hematopoietic progenitors at various stages of differentiation and development of normal myeloid cells [2,3,4]. The incidence of AML increased proportionally with age, from 1.8 cases per 100,000 people under 65 years old to 13.7 cases per 100,000 people over 65 years old. In developed countries, various half of novelly diagnosed AML sufferers are over 65 years, with a median age of 67 years at diagnosis, and more men have AML than women [5]. There are various 20,000 new AML cases in the US in 2021 [6]. On the other hand, merely about 25% of AML sufferers survived for 5 years or more despite multi-drug combination chemotherapy. Elderly sufferers (> 60 years) and sufferers that could not bear standard induction chemotherapy had unfavorable molecular outcomes, with a median survival of merely 5–10 months and a 5-year overall survival (OS) of 5% [7, 8]. At present, scientific research has moved from the cellular level to the molecular level. For example, targeted drugs [9], nanomaterials [10], and molecular markers [11]. We are committed to finding new molecular markers to more accurately predict the progress and prognosis of AML.
Microchromosome maintenance protein 5 (MCM5) is a key cell cycle regulator located on chromosome 22Q13.1 [12], whose role in DNA replication is verified [13]. MCM5 is a DNA licensing factor as an ingredient of the MCM2-7 complex, a putative replicative helicase vital for the initiation and extension of "once per cell cycle" DNA replication in eukaryotic cells [14, 15]. So far, MCM5 has been reported to be closely correlated with varieties of diseases. For instance, increased level of MCM5 is remarkably correlated with the positive progression and unfavorable prognosis of oral squamous cell carcinoma, and MCM5 can be utilized as a marker for the early diagnosis of oral squamous cell carcinoma [16, 17]. MCM5 was also reported to be an individual prognostic element in lung squamous cell carcinoma [18]. MCM5 can aggravate the HDAC1-mediated malignant progression of lung cancer [19]. MCM5 is correlated with malignant status and unfavorable prognosis in cervical adenocarcinoma sufferers, and regulates the proliferation of cervical adenocarcinoma cells [20]. MCM5 is a new sensitive as well as specific biomarker for the detection of endometrial and ovarian tumors in urine samples [21]. Therefore, we investigated the relationship betwixt MCM5 and AML.
In this study, we investigated the gene level microarray of AML and correlative clinical data in GSE38865, GSE142698, and The Cancer Genome Atlas (TCGA) database. MCM5 was selected as the research objective. Then, we studied the level profile and biological functions of MCM5 in AML, and further analyzed the correlation betwixt MCM5 level and the AML.
Methods
Data acquisition
With gene expression omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database, “AML” and “survival” in the search box as the keyword, we harvested the AML gene level chips and correlative clinical data. GSE38865 [22] was selected as the training data set of this study. We extracted clinical data with prognostic information directly from the matrix file on the correlative gene chip page in the GEO database.
From TCGA (https://portal.gdc.cancer.gov/) we harvested RNA-seq data (TPM) and the correlative clinical information of AML.
Screening of Prognosis-Related Genes
Kaplan–Meier curves and Cox regression analysis to screen genes correlated to survival. The prognosis-related genes screened through “survival” R package with the KM < 0.01 and cox p value < 0.01 were shown in Table S1.
Identify Target Gene
After screening of prognosis-related genes, we utilized multivariate Cox regression analysis to perform individual prognostic analysis (p < 0.01). Thus, we got the genes remarkably correlated with individual prognosis which were exhibited in Table 1.
Next, we analyze the correlation betwixt individual prognostic-related genes and clinical traits through Wilcox.test or Kruskal.test (p < 0.05). The correlation betwixt individual prognostic-related genes and clinical traits was shown in Table 2.
Then, we utilized GEPIA (http://gepia2.cancer-pku.cn) to explore the relationship betwixt genes in Table 2 (SigNum > 0) and the survival of AML.GEPIA is an interactive web server [23].
Survival Analysis
All AML sufferers with MCM5 level values higher than the median were classified as MCM5 high group, and the remaining sufferers were classified as MCM5 low group. We utilized Kaplan–Meier curve and log-rank test via the R package “survival” and “survminer” to analyze the survival of MCM5 in the prognostic model. Individual prognostic value of MCM5 was verified with multivariate and univariate Cox regression analyses.
Enrichment Analysis
The difference of gene level in AML samples betwixt MCM5 high group and MCM5 low group was compared through setting adjusted p < 0.05 and fold change > 1 threshold using the ‘‘Limma” package. "ClusterProfiler" and "enrichplot" packages were utilized for Kyoto encyclopedia of genes and genomes (KEGG) analysis and Gene Ontology (GO) to further explore the functions of various genes.
Statistical Analysis
Through R software 3.5.0 we performed all statistical analysis. We utilized Fisher exact test as well as the Wilcoxon rank-sum tests, respectively, to verify hypotheses for categorical and continuous variables. According to the median level value of MCM5, the samples in the second cohort were divided into MCM5 high group (n = 10) and MCM5 low group (n = 10). The limma package was utilized to analyze the distinct gene level. Kaplan–Meier method and Cox regression multivariate analysis were utilized for survival analysis, and log-rank test was utilized for comparison betwixt groups. To identify GO and KEGG enrichment terms, we utilized “Clusterprofiler” package. For all statistical analysis, p < 0.05 was considered significant.
Results
Screening Target Genes
We got GSE38865 from GEO database (https://www.ncbi.nlm.nih.gov/geo/). According to the probe information of Illumina HumanWG-6 V3.0 expression beadchip (GPL6884) and Illumina HumanHT-12 V4.0 expression beadchip (GPL10558), 22089 genes of 30 AML samples were annotated. Then, we performed Kaplan–Meier curves and Cox regression analysis to screen genes correlated to survival. The prognosis-related genes were screened through “survival” R package with the KM < 0.01 and coxPvalue < 0.01 (Table S1).
In order to get the genes for independent prognostic, multivariate Cox regression analysis was performed (p < 0.01) in prognosis-related genes. 52 genes for independent prognostic were exhibited in Table 1.
Later, we analyze the correlation betwixt individual prognostic genes and clinical traits (age, gender, Wbc, and NPM1 mutation) through wilcox.test or kruskal.test (p < 0.05). Four clinical traits related-individual prognostic genes (DENND4C, DKC1, LAPTM4B, and MCM5) were shown in Table 2.
GEPIA (http://gepia2.cancer-pku.cn) was utilized to verify the relationship betwixt these 4 genes and the over survival of AML. We found that merely MCM5 was correlated to the survival and prognosis of AML. As presented in Fig. 1, sufferers with high level of MCM5 had worse prognosis than these with low level of MCM5. Therefore, we chose MCM5 for follow-up study.
Identify Target Gene MCM5
In GSE38865, all AML sufferers with MCM5 level values above the median were classified as MCM5 high group, and the others were considered to be MCM5 low group. As shown in Fig. 2A, the prognosis of AML sufferers with high MCM5 level was worse than that of sufferers with low MCM5 level. Multivariate and univariate Cox regression analysis exhibited that MCM5 was remarkably correlated with the prognosis of AML sufferers and was an independent prognostic element. (Fig. 2B and C; Table S2).
MCM5 in TCGA database
To further identify target gene MCM5, we harvested RNA-seq data (TPM) and the correlative clinical information from TCGA-AML (https://portal.gdc.cancer.gov/).
We analyzed these MCM5 level based on the TCGA-AML. The results were consistent with those harvested from GSE38865 data. In Fig. 3, high MCM5 group had worse prognosis than low MCM5 group (Fig. 3A), and MCM5 was an independent prognostic element in AML (Fig. 3B and C).
MCM5 Overexpression in AML
Then, we want to study whether MCM5 level shows difference in AML and normal samples. GSE142698 (AML = 24; normal = 24) was download from GEO database. We found an increase level of MCM5 in AML blood compared with healthy blood (Fig. 4A). We also found that an increase level in AML bone marrow samples compared with T Acute Lymphoblastic Leukemia (T-ALL) bone marrow samples (Fig. 4B) based on GSE131184 (AML = 76; T-ALL = 49).
Differences in Clinical in AML Betwixt MCM5 High Group and MCM5 Low Groups
The differences in clinical in AML betwixt MCM5 high group and MCM5 low groups were analyzed through “ggpubr” package. There was no significant difference in the age gender, and NPM1 mutation betwixt MCM5 high group and MCM5 low groups (Fig. S1A-C).
Different Expression and Functional Enrichment Analysis for Genes in MCM5 High Group Versus MCM5 Low Group
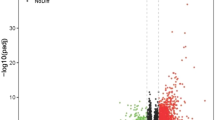
To discover the genes correlated with MCM5, we analyzed the discrepant level values betwixt MCM5 high group and MCM5 low group (adj.p < 0.05, (FC, log2) > 1 or < 1). There were 34 up-modulated genes and 47 down-modulated genes (Fig. 5A; Table S3). Figure 5B exhibited the top 20 genes in up-modulated and down-modulated groups. We analyzed the enriched GO terms through using the discrepantly expressed genes (DEGs). Among the biological process terms of GO, DEGs are mainly enriched in regulating extracellular matrix organization, negatively regulating response to external stimulus, phase transition of mitotic cell cycle, regulating mitotic cell cycle, and DNA replication (Fig. 5C). Showed by the KEGG analysis results, DNA replication, Kaposi sarcoma-correlated herpesvirus infection, Cell cycle, Glycine, serine and threonine metabolism, and Amino sugar and nucleotide sugar metabolism were the most enriched pathways (Fig. 5D).
Module Screening Using PPI Network
Last, the correlation analysis of the top 40 DEGs betwixt MCM5 high group and MCM5 low group was performed (Fig. 6A). We could see that MCM5 was positive correlation with KIAA0101 which also up-modulated in MCM5 high group.
KIAA0101 is a proliferating cell nuclear antigen (PCNA) -related element. KIAA0101 has been verified to be over-expressed in a variety of human malignant tumors, such as esophageal squamous cell carcinoma, lung adenocarcinoma, chronic lymphocytic leukemia, and it has been identified as an oncogene [24,25,26,27,28]. Protein–protein interaction (PPI) network was screened in the String database through with the 81 DEGs. All down-modulated genes and most up-modulated genes had interactions in the PPI network (Fig. 6B). In the general network MCM5 acts as a core gene. The graphical abstract is showed in Fig. S2.
Discussion
AML is listed among the most common hematologic malignancies [29, 30]. Despite progress in the treatment of AML, it remains difficult to treat and cure, and the 5-year overall survival rate, especially in sufferers which age is over 60 years, is still very low [31,32,33]. The pathogenesis, diagnosis and treatment of AML are hot topics in current research.
In our research, we analyzed the mRNA and the correlative clinical information in GSE38865. We wanted to find the genes which could predict bad or good prognosis in AML. Through using Kaplan–Meier curves, Cox regression analysis, Wilcox.test or Kruskal.test, and GEPIA, we found MCM5 may the target gene which was an independent prognostic element for AML. From TCGA we also downloaded the AML-related RNA-seq data (TPM) and harvested the correlative clinical information. It was exhibited by results that MCM5 was an independent prognostic element for AML and AML sufferers in MCM5 high group had worse prognosis than that in MCM5 low group. We also found an increase level in AML blood compared with healthy blood (GSE142698) and an increase level in AML bone marrow samples compared with T-ALL bone marrow samples (GSE131184).
MCM5, as a critical cell cycle regulator and DNA replication licensing element, was expressed highly in multiple cancer tissues, such as cervical adenocarcinoma, cervical cancer, renal cell carcinoma, and laryngeal squamous cell cancer [20, 34,35,36]. MCM5 in thyroid cancer cells was also reported as a target of BET inhibitors. MCM5 may serve as an adverse prognostic biomarker for lung cancer[18, 37, 38]. In addition, Chen Liu even reported that compared with leukocytes, MCM5 was highly expressed in AML cell lines (KG-1a, NB4 and HL60)[12].
To find the genes correlated with MCM5, we analyzed the deferent expressed genes betwixt MCM5 high group and MCM5 low group. We found CCL1, MLLT11, and PIM1 were up-modulated in MCM5 high group. CCL1 is involved in immune-cell recruitment and, like other chemokines, is involved in nociceptive processing[39]. The chemokine CCL1 activates the AMFR-SPRY1 pathway, which facilitates differentiation of pulmonary fibroblasts into myofibroblasts and drives pulmonary fibrosis[40]. MLLT11 acted as an oncogene in multiple cancers, for example osteosarcoma[41], bladder cancer[42], and lung cancer[43]. Importantly, report exhibited that MLLT11 is a unfavorable prognostic biomarker for AML, adult normal cytogenetics AML, and adult myelodysplastic syndrome[44]. PIM1 plays a critical role in the development of many hematopoietic and non-hematopoietic malignancies, including prostate cancer and acute myeloid leukemia[45, 46]. AML with high level of PIM1 was reported to show an unfavorable prognosis. PIM1 facilitates the proliferation and inhibits apoptosis of AML cells, but also enhances the chemotactic ability of leukemia cells[47, 48].
Likewise, the GO pathways mainly enriched in regulation of extracellular matrix organization, DNA replication, mitotic cell cycle phase transition, regulation of mitotic cell cycle, mitotic cell cycle phase transition, and negative regulation of response to external stimulus. DNA replication, Kaposi sarcoma-correlated herpesvirus infection, Cell cycle, Amino sugar and nucleotide sugar metabolism, serine and threonine metabolism, and Glycine.
PPI network also revealed that MCM5 is highly linked with KIAA0101. KIAA0101 is a proliferating cell nuclear antigen (PCNA) -related element. KIAA0101 has been reported to be over-expressed in many human malignant tumors and has been identified as an oncogene[27, 49]
Conclusions
In conclusion, we demonstrated that MCM5 was an independent prognostic element for AML. Low level of MCM5 was a good prognostic element for AML sufferers. In addition, the results of GO term enrichment, KEGG analysis and PPI network involvement in AML showed that MCM5 may regulate DNA replication and cell cycle of AML cells which provided an insight into the pathogenesis correlated with different MCM5 level. However, the exact pathophysiological role of MCM5 in AML cells has not been fully demonstrated in this study. Further investigation of the molecular mechanism of MCM5 in AML progression and more in-depth genomic studies are urgently needed.
References
Pirillo, C., Birch, F., Tissot, F. S., Anton, S. G., Haltalli, M., Tini, V., Kong, I., Piot, C., Partridge, B., Pospori, C., Keeshan, K., Santamaria, S., Hawkins, E., et al. (2022). Metalloproteinase inhibition reduces AML growth, prevents stem cell loss, and improves chemotherapy effectiveness. Blood Advances, 6(10), 3126–3141.
Arber, D. A., Orazi, A., Hasserjian, R., Thiele, J., Borowitz, M. J., Le Beau, M. M., Bloomfield, C. D., Cazzola, M., & Vardiman, J. W. (2016). The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood, 127(20), 2391–2405.
Shang, Z., Ming, X., Wu, J., & Xiao, Y. (2021). Downregulation of circ_0012152 inhibits proliferation and induces apoptosis in acute myeloid leukemia cells through the miR-625-5p/SOX12 axis. Hematological Oncology, 39(4), 539–548.
Ben Khoud, M., Ingegnere, T., Quesnel, B., Mitra, S., & Brinster, C. (2021). Acute myeloid leukemia: is it T time? Cancers, 13(10), 2385.
Fey MF, Buske C and Group EGW. Acute myeloblastic leukaemias in adult patients: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013; 24 Suppl 6:vi138–143.
Bewersdorf, J. P., & Abdel-Wahab, O. (2022). Translating recent advances in the pathogenesis of acute myeloid leukemia to the clinic. Genes & Development, 36(5–6), 259–277.
Zhang, Y., Xue, S., Hao, Q., Liu, F., Huang, W., & Wang, J. (2021). Galectin-9 and PSMB8 overexpression predict unfavorable prognosis in patients with AML. Journal of Cancer, 12(14), 4257–4263.
Lai, C., Doucette, K., & Norsworthy, K. (2019). Recent drug approvals for acute myeloid leukemia. Journal of Hematology & Oncology, 12(1), 100.
Ashique, S., Sandhu, N. K., Chawla, V., & Chawla, P. A. (2021). Targeted drug delivery: Trends and perspectives. Current Drug Delivery, 18(10), 1435–1455.
Yousefi, S. R., Sobhani, A., Alshamsi, H. A., & Salavati-Niasari, M. (2021). Green sonochemical synthesis of BaDy2NiO5/Dy2O3 and BaDy2NiO5/NiO nanocomposites in the presence of core almond as a capping agent and their application as photocatalysts for the removal of organic dyes in water. RSC Advances, 11(19), 11500–11512.
Giordano, C., Karasik, O., King-Morris, K., & Asmar, A. (2015). Uric acid as a marker of kidney disease: Review of the current literature. Disease Markers, 2015, 382918.
Liu, C., Zhong, L., Shen, C., Chu, X., Luo, X., Yu, L., Ye, J., Xiong, L., Dan, W., Li, J., & Liu, B. (2021). CRNDE enhances the expression of MCM5 and proliferation in acute myeloid leukemia KG-1a cells by sponging miR-136-5p. Science and Reports, 11(1), 16755.
Zhang, Y., Xia, J., Liu, M., Chen, B., Yang, M., Yu, X., Ou, Y., Li, S., Liu, X., Feng, Y., Su, B., & Huang, S. (2022). Mcm5 represses endodermal migration through Cxcr4a-itgb1b cascade instead of cell cycle control. Biomolecules, 12(2), 286.
Vetro, A., Savasta, S., Russo Raucci, A., Cerqua, C., Sartori, G., Limongelli, I., Forlino, A., Maruelli, S., Perucca, P., Vergani, D., Mazzini, G., Mattevi, A., Stivala, L. A., et al. (2017). MCM5: A new actor in the link between DNA replication and Meier-Gorlin syndrome. European Journal of Human Genetics, 25(5), 646–650.
Burger, M. (2009). MCM2 and MCM5 as prognostic markers in colon cancer: A worthwhile approach. Digestive Diseases and Sciences, 54(2), 197–198.
Yu, S. Y., Wang, Y. P., Chang, J. Y., Shen, W. R., Chen, H. M., & Chiang, C. P. (2014). Increased expression of MCM5 is significantly associated with aggressive progression and poor prognosis of oral squamous cell carcinoma. Journal of Oral Pathology and Medicine, 43(5), 344–349.
Hao, M., Wang, H., Zhang, C., Li, C., & Wang, X. (2020). Minichromosome maintenance protein 5 is an important pathogenic factor of oral squamous cell carcinoma. Oncology Letters, 20(4), 109.
Liu, Y. Z., Wang, B. S., Jiang, Y. Y., Cao, J., Hao, J. J., Zhang, Y., Xu, X., Cai, Y., & Wang, M. R. (2017). MCMs expression in lung cancer: Implication of prognostic significance. Journal of Cancer, 8(18), 3641–3647.
Zhang, L. L., Li, Q., Zhong, D. S., Zhang, W. J., Sun, X. J., & Zhu, Y. (2021). MCM5 aggravates the HDAC1-mediated malignant progression of lung cancer. Frontiers in Cell and Developmental Biology, 9, 669132.
Wang, D., Li, Q., Li, Y., & Wang, H. (2018). The role of MCM5 expression in cervical cancer: Correlation with progression and prognosis. Biomedicine & Pharmacotherapy, 98, 165–172.
Stockley, J., Akhand, R., Kennedy, A., Nyberg, C., Crosbie, E. J., & Edmondson, R. J. (2020). Detection of MCM5 as a novel non-invasive aid for the diagnosis of endometrial and ovarian tumours. BMC Cancer, 20(1), 1000.
Diffner, E., Beck, D., Gudgin, E., Thoms, J. A., Knezevic, K., Pridans, C., Foster, S., Goode, D., Lim, W. K., Boelen, L., Metzeler, K. H., Micklem, G., Bohlander, S. K., et al. (2013). Activity of a heptad of transcription factors is associated with stem cell programs and clinical outcome in acute myeloid leukemia. Blood, 121(12), 2289–2300.
Tang, Z., Li, C., Kang, B., Gao, G., Li, C., & Zhang, Z. (2017). GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research, 45(W1), W98–W102.
Sun, Z., Zhang, S., Zhang, N., Wang, J., Wang, J., & Liu, J. (2022). Circ_0005231 promotes the progression of esophageal squamous cell carcinoma via sponging miR-383-5p and regulating KIAA0101. Thoracic Cancer, 13(12), 1751–1762.
Zhou, B., Zang, R., Zhang, M., Song, P., Liu, L., Bie, F., Peng, Y., Bai, G., & Gao, S. (2022). Identifying novel tumor-related antigens and immune phenotypes for developing mRNA vaccines in lung adenocarcinoma. International Immunopharmacology, 109, 108816.
Deng, Y., Chen, X., Huang, C., Song, J., Feng, S., Chen, X., & Zhou, R. (2022). Screening and validation of significant genes with poor prognosis in pathologic Stage-I lung adenocarcinoma. Journal of Oncology, 2022, 3794021.
Wang, K., Li, J., & Zhou, B. (2022). KIAA0101 knockdown inhibits glioma progression and glycolysis by inactivating the PI3K/AKT/mTOR pathway. Metabolic Brain Disease, 37(2), 489–499.
Zhang, Q., Yuan, J., Liu, Y., Liu, X., Lv, T., Zhou, K., & Song, Y. (2021). KIAA0101 knockdown inhibits cell proliferation and induces cell cycle arrest and cell apoptosis in chronic lymphocytic leukemia cells. Annals of Translational Medicine, 9(6), 487.
Bhattacharjee, R., Ghosh, S., Nath, A., Basu, A., Biswas, O., Patil, C. R., & Kundu, C. N. (2022). Theragnostic strategies harnessing the self-renewal pathways of stem-like cells in the acute myeloid leukemia. Critical Reviews in Oncology Hematology, 177, 103753.
Liu, G., Zhang, Q., Yang, J., Li, X., Xian, L., Li, W., Lin, T., Cheng, J., Lin, Q., Xu, X., Li, Q., Lin, Y., Zhou, M., et al. (2022). Increased TIGIT expressing NK cells with dysfunctional phenotype in AML patients correlated with poor prognosis. Cancer Immunology, Immunotherapy, 71(2), 277–287.
Liu, L., Patnana, P. K., Xie, X., Frank, D., Nimmagadda, S. C., Su, M., Zhang, D., Koenig, T., Rosenbauer, F., Liebmann, M., Klotz, L., Xu, W., Vorwerk, J., et al. (2022). GFI1B acts as a metabolic regulator in hematopoiesis and acute myeloid leukemia. Leukemia, 36(9), 2196–2207.
Zhang, F., Sun, J., Tang, X., Liang, Y., Jiao, Q., Yu, B., Dai, Z., Yuan, X., Li, J., Yan, J., Zhang, Z., Fan, S., Wang, M., et al. (2022). Stabilization of SAMHD1 by NONO is crucial for Ara-C resistance in AML. Cell Death & Disease, 13(7), 590.
Trad, R., Warda, W., Alcazer, V., Neto da Rocha, M., Berceanu, A., Nicod, C., Haderbache, R., Roussel, X., Desbrosses, Y., Daguindau, E., Renosi, F., Roumier, C., Bouquet, L., et al. (2022). Chimeric antigen receptor T-cells targeting IL-1RAP: a promising new cellular immunotherapy to treat acute myeloid leukemia. Journal of Immunotherapy Cancer, 10(7).
Wang, D., Wang, H., Li, Y., & Li, Q. (2018). MiR-362-3p functions as a tumor suppressor through targeting MCM5 in cervical adenocarcinoma. Bioscience Report 38(3)
Nowinska, K., Ciesielska, U., Piotrowska, A., Jablonska, K., Partynska, A., Paprocka, M., Zatonski, T., Podhorska-Okolow, M., & Dziegiel, P. (2019). MCM5 expression is associated with the grade of malignancy and Ki-67 antigen in LSCC. Anticancer Research, 39(5), 2325–2335.
Gong, B., Ma, M., Yang, X., Xie, W., Luo, Y., & Sun, T. (2019). MCM5 promotes tumour proliferation and correlates with the progression and prognosis of renal cell carcinoma. International Urology and Nephrology, 51(9), 1517–1526.
Sun, M., Wang, T., Zhu, Y., Zhang, Y., Zhu, L., & Li, X. (2022). The high expression of minichromosome maintenance complex component 5 is an adverse prognostic factor in lung adenocarcinoma. BioMed Research International, 2022, 4338793.
Liu, K., Kang, M., Liao, X., & Wang, R. (2019). Genome-wide investigation of the clinical significance and prospective molecular mechanism of minichromosome maintenance protein family genes in patients with Lung Adenocarcinoma. PLoS ONE, 14(7), e0219467.
García-Domínguez, M., González-Rodríguez, S., Hidalgo, A., Baamonde, A., & Menéndez, L. (2021). Kappa-opioid receptor-mediated thermal analgesia evoked by the intrathecal administration of the chemokine CCL1 in mice. Fundamental & Clinical Pharmacology, 35(6), 1109–1118.
Liu, S. S., Liu, C., Lv, X. X., Cui, B., Yan, J., Li, Y. X., Li, K., Hua, F., Zhang, X. W., Yu, J. J., Yu, J. M., Wang, F., Shang, S., et al. (2021). The chemokine CCL1 triggers an AMFR-SPRY1 pathway that promotes differentiation of lung fibroblasts into myofibroblasts and drives pulmonary fibrosis. Immunity, 54(9), 2042–2056.
Man, G., Duan, A., Liu, W., Cheng, J., Liu, Y., Song, J., Zhou, H., & Shen, K. (2021). Circular RNA-related CeRNA network and prognostic signature for patients with osteosarcoma. Cancer Management Research, 13, 7527–7541.
Jin, H., Sun, W., Zhang, Y., Yan, H., Liufu, H., Wang, S., Chen, C., Gu, J., Hua, X., Zhou, L., Jiang, G., Rao, D., Xie, Q., et al. (2018). MicroRNA-411 downregulation enhances tumor growth by upregulating MLLT11 expression in human bladder cancer. Mol Ther Nucleic Acids, 11, 312–322.
Zhang, H. P., Chen, Q. K., & Xu, J. F. (2020). LPAR5 stimulates the malignant progression of non-small-cell lung carcinoma by upregulating MLLT11. European Review for Medical and Pharmacological Sciences, 24(17), 8902–8910.
Xiong, Y., Li, Z., Ji, M., Tan, A. C., Bemis, J., Tse, J. V., Huang, G., Park, J., Ji, C., Chen, J., Bemis, L. T., Bunting, K. D., & Tse, W. (2011). MIR29B regulates expression of MLLT11 (AF1Q), an MLL fusion partner, and low MIR29B expression associates with adverse cytogenetics and poor overall survival in AML. British Journal of Haematology, 153(6), 753–757.
Anjum, F., Mohammad, T., Almalki, A. A., Akhtar, O., Abdullaev, B., & Hassan, M. I. (2021). Phytoconstituents and medicinal plants for anticancer drug discovery: Computational identification of potent inhibitors of PIM1 kinase. OMICS, 25(9), 580–590.
Takeuchi, H., Miyamoto, T., Fuseya, C., Asaka, R., Ida, K., Ono, M., Tanaka, Y., Shinagawa, M., Ando, H., Asaka, S., & Shiozawa, T. (2022). PIM1 is a poor prognostic factor for and potential therapeutic target in serous carcinoma of the endometrium. International Journal of Gynecological Pathology. https://doi.org/10.1097/PGP.0000000000000882
Li, Q., Sun, R. X., Ou, Y., Luo, H. M., & Wu, Y. (2019). Role of Pim1 gene overexpression in pathogenesis of acute myeloid leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi, 27(3), 664–672.
Goldberg, L., Tijssen, M. R., Birger, Y., Hannah, R. L., Kinston, S. J., Schutte, J., Beck, D., Knezevic, K., Schiby, G., Jacob-Hirsch, J., Biran, A., Kloog, Y., Marcucci, G., et al. (2013). Genome-scale expression and transcription factor binding profiles reveal therapeutic targets in transgenic ERG myeloid leukemia. Blood, 122(15), 2694–2703.
Lin, P., Zhao, Y., Li, X., & Liang, Z. (2021). KIAA0101 in Malignant Pleural Mesothelioma: a Potential Diagnostic and Prognostic Marker. Comb Chem High Throughput Screen.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, S., Wu, W. & Han, X. Enhanced MCM5 Level Predicts Bad Prognosis in Acute Myeloid Leukemia. Mol Biotechnol 65, 1242–1252 (2023). https://doi.org/10.1007/s12033-022-00623-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00623-9