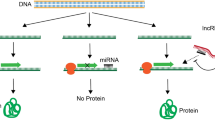
Abstract
As reported by WHO in 2018, there were 2.09 million victims of lung cancer and 1.76 million fatalities worldwide. Tobacco remains the biggest hazard in causing this lethal disease. To execute the computational analysis, the overexpressed lung cancer genes were retrieved from literature and subsequently their complete coding sequences (CDS) were downloaded. The mature microRNA sequences of human were extracted from miRBASE. The 7mer-m8 perfect seed match between miRNAs and mRNAs was found. Following filtration, 7 genes were selected that possessed binding sites for maximum miRNAs viz., MUC5B (miR-4479, miR-1227-5p, miR-3940-5p, miR-604, miR-4455, miR-4267, miR-6750-3p, miR-4530, miR-5587-5p, miR-4508, miR-4534, miR-4443, miR-4253, miR-1321, miR-4655-5p, miR-4297, miR-4296, miR-1268a, miR-3178, miR-4750-3p, miR-1306-3p, miR-1268b, miR-3656, miR-1233-3p, miR-6804-5p), MUC16 (miR-4456, miR-1205, miR-665, miR-6808-3p, miR-1279, miR-4257, miR-1227-5p, miR-888-3p, miR-4455, miR-4267, miR-4294, miR-1275, miR-4288, miR-1178-5p, miR-4314, miR-6829-3p, miR-548av-5p, miR-1294, miR-5587-5p, miR-3622b-5p, miR-1273f, miR-4770, miR-4327, miR-4318, miR-4531, miR-4534, miR-4443, miR-7106-5p, miR-3125, miR-3650, miR-4325, miR-4266, miR-7976, miR-1290, miR-4500, miR-7160-5p, miR-4291, miR-1306-3p, miR-6130, miR-4430, miR-4725-5p, miR-4441, miR-6077, miR-1304-5p, miR-7515, miR-3182, miR-6134), COL1A1 (miR-3665, miR-1227-5p, miR-6132, miR-2861, miR-4530, miR-3155b, miR-3155a, miR-1292-3p, miR-4497), COL5A1 (miR-7162-5p, miR-3665, miR-6809-3p, miR-4313, miR-4531, miR-4532, miR-3155b, miR-4323, miR-1207-3p, miR-4260, miR-6071, miR-4710, miR-7162-5p), CELSR2 (miR-7150, miR-4308, miR-6132, miR-4770, miR-4534, miR-4492, miR-3960, miR-3178, miR-4291, miR-563), COL7A1 (miR-665, miR-6730-3p, miR-1227-5p, miR-4265, miR-6829-3p, miR-4297, miR-4532, miR-3181, miR-4310, miR-4441, miR-4497, miR-1237-3p), and FAT2 (miR-4267, miR-1275, miR-4770, miR-1825, miR-6895-5p, miR-4535, miR-4493, miR-940, miR-6861-3p, miR-4310, miR-4710, miR-4447, miR-4472). The miRNA-target site and their flank regions were compared with respect to site accessibility, translational rate, and relationship between RSCU and tRNAs. Higher accessibilities to miRNA-binding regions and lower translational rates indicated that miRNAs’ binding to their respective targets might be efficient. The presence of rare codons might further augment miRNA targeting. The codon usage bias study of the genes related to lung cancer revealed non-uniform usage of nucleotides and comparatively higher GC content. Lower biasness prevailed in the genes and selective constraint mostly governed them. Lastly, the functionalities of target genes were also revealed. The silencing characteristic of miRNAs might be exploited to design miRNA-mediated therapy that might potentially repress the overexpressed genes in carcinoma.




Similar content being viewed by others
Data Availability
All data are available in the manuscript and in the supplementary file.
Code Availability
Not applicable.
References
Lemjabbar-Alaoui, H., Hassan, O. U., Yang, Y.-W., & Buchanan, P. (2015). Lung cancer: Biology and treatment options. Biochimica et Biophysica Acta (BBA), 1856(2), 189–210.
Cruz, C. S. D., Tanoue, L. T., & Matthay, R. A. (2011). Lung cancer: Epidemiology, etiology, and prevention. Clinics in Chest Medicine, 32(4), 605–644.
Bartel, D. P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell, 136(2), 215–233.
Kim, D., Chang, H. R., & Baek, D. (2017). Rules for functional microRNA targeting. BMB reports, 50(11), 554.
Shen, Z., Zhang, Y.-H., Han, K., Nandi, A. K., Honig, B., & Huang, D.-S. (2017). miRNA-disease association prediction with collaborative matrix factorization. Complexity, 2017, 2498957. https://doi.org/10.1155/2017/2498957
Rupaimoole, R., & Slack, F. J. (2017). MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nature reviews Drug discovery, 16(3), 203.
Hanna, J., Hossain, G. S., & Kocerha, J. (2019). The Potential for microRNA Therapeutics and Clinical Research. Frontiers in Genetics. https://doi.org/10.3389/fgene.2019.00478
Mognato, M., & Celotti, L. (2015). MicroRNAs used in combination with anti-cancer treatments can enhance therapy efficacy. Mini reviews in medicinal chemistry, 15(13), 1052–1062.
Si, W., Shen, J., Zheng, H., & Fan, W. (2019). The role and mechanisms of action of microRNAs in cancer drug resistance. Clinical epigenetics, 11(1), 25.
Reczko, M., Maragkakis, M., Alexiou, P., Grosse, I., & Hatzigeorgiou, A. G. (2012). Functional microRNA targets in protein coding sequences. Bioinformatics, 28(6), 771–776.
Zhang, K., Zhang, X., Cai, Z., Zhou, J., Cao, R., Zhao, Y., & Zhao, Q. (2018). A novel class of microRNA-recognition elements that function only within open reading frames. Nature Structural & Molecular Biology, 25(11), 1019–1027.
Gu, W., Wang, X., Zhai, C., Zhou, T., & Xie, X. (2013). Biological basis of miRNA action when their targets are located in human protein coding region. PLoS ONE, 8(5), e63403.
Peterson, S., Thompson, J., Ufkin, M., Sathyanarayana, P., Liaw, L., & Congdon, C. B. (2014). Common features of microRNA target prediction tools. Frontiers in Genetics. https://doi.org/10.3389/fgene.2014.00023
Liu, W., & Wang, X. (2019). Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biology, 20(1), 18. https://doi.org/10.1186/s13059-019-1629-z
Gu, W., Zhai, C., Wang, X., Xie, X., Parinandi, G., & Zhou, T. (2012). Translation efficiency in upstream region of microRNA targets in Arabidopsis thaliana. Evolutionary Bioinformatics, 8, 10362.
Gu, W., Wang, X., Zhai, C., Xie, X., & Zhou, T. (2012). Selection on synonymous sites for increased accessibility around miRNA binding sites in plants. Molecular Biology and Evolution, 29(10), 3037–3044.
Athey, J., Alexaki, A., Osipova, E., Rostovtsev, A., Santana-Quintero, L. V., Katneni, U., & Kimchi-Sarfaty, C. (2017). A new and updated resource for codon usage tables. BMC Bioinformatics, 18(1), 391. https://doi.org/10.1186/s12859-017-1793-7
Guan, D.-L., Ma, L.-B., Khan, M. S., Zhang, X.-X., Xu, S.-Q., & Xie, J.-Y. (2018). Analysis of codon usage patterns in Hirudinaria manillensis reveals a preference for GC-ending codons caused by dominant selection constraints. BMC Genomics, 19(1), 542. https://doi.org/10.1186/s12864-018-4937-x
Gun, L., Yumiao, R., Haixian, P., & Liang, Z. (2018). Comprehensive Analysis and Comparison on the Codon Usage Pattern of Whole<i> Mycobacterium tuberculosis</i> Coding Genome from Different Area. BioMed Research International, 2018, 3574976. https://doi.org/10.1155/2018/3574976
Song, H., Liu, J., Song, Q., Zhang, Q., Tian, P., & Nan, Z. (2017). Comprehensive analysis of codon usage bias in seven epichloë species and their peramine-coding genes. Frontiers in microbiology. https://doi.org/10.3389/fmicb.2017.01419
LaBella, A. L., Opulente, D. A., Steenwyk, J. L., Hittinger, C. T., & Rokas, A. (2019). Variation and selection on codon usage bias across an entire subphylum. PLoS Genetics, 15(7), e1008304. https://doi.org/10.1371/journal.pgen.1008304
Plotkin, J. B., Robins, H., & Levine, A. J. (2004). Tissue-specific codon usage and the expression of human genes. Proceedings of the National Academy of Sciences of the United States of America, 101(34), 12588–12591. https://doi.org/10.1073/pnas.0404957101
Axelsen, J. B., Lotem, J., Sachs, L., & Domany, E. (2007). Genes overexpressed in different human solid cancers exhibit different tissue-specific expression profiles. Proceedings of the National Academy of Sciences, 104(32), 13122–13127. https://doi.org/10.1073/pnas.0705824104
Lewis, B. P., Shih, I.-H., Jones-Rhoades, M. W., Bartel, D. P., & Burge, C. B. (2003). Prediction of mammalian microRNA targets. Cell, 115(7), 787–798.
Kertesz, M., Iovino, N., Unnerstall, U., Gaul, U., & Segal, E. (2007). The role of site accessibility in microRNA target recognition. Nature Genetics, 39(10), 1278.
Hausser, J., Landthaler, M., Jaskiewicz, L., Gaidatzis, D., & Zavolan, M. (2009). Relative contribution of sequence and structure features to the mRNA binding of Argonaute/EIF2C–miRNA complexes and the degradation of miRNA targets. Genome Research, 19(11), 2009–2020.
SantaLucia, J. (1998). A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proceedings of the National Academy of Sciences, 95(4), 1460–1465.
Tuller, T., Waldman, Y. Y., Kupiec, M., & Ruppin, E. (2010). Translation efficiency is determined by both codon bias and folding energy. Proceedings of the National Academy of Sciences of the United States of America, 107(8), 3645–3650. https://doi.org/10.1073/pnas.0909910107
Dilucca, M., Cimini, G., Semmoloni, A., Deiana, A., & Giansanti, A. (2015). Codon bias patterns of E. coli’s interacting proteins. PLoS ONE, 10(11), e0142127.
Sun, S., Xiao, J., Zhang, H., & Zhang, Z. (2016). Pangenome evidence for higher codon usage bias and stronger translational selection in core genes of E. coli. Frontiers in Microbiology, 7, 1180. https://doi.org/10.3389/fmicb.2016.01180
Sharp, P. M., & Li, W.-H. (1986). Codon usage in regulatory genes in Escherichia coli does not reflect selection for ‘rare’codons. Nucleic Acids Research, 14(19), 7737–7749.
Wan, X., Xu, D., & Zhou, J. (2003). A new informatics method for measuring synonymous codon usage bias. Intelligent engineering systems through artificial neural networks 13.
Yao, H., Chen, M., & Tang, Z. (2019). Analysis of synonymous codon usage bias in flaviviridae virus. BioMed Research International, 2019, 5857285. https://doi.org/10.1155/2019/5857285
Conesa, A., & Gotz, S. (2008). Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics, 2008, 619832. https://doi.org/10.1155/2008/619832
Dong, F., Ji, Z.-B., Chen, C.-X., Wang, G.-Z., & Wang, J.-M. (2013). Target gene and function prediction of differentially expressed microRNAs in lactating mammary glands of dairy goats. International Journal of Genomics, 2013, 1–13.
Dai, X., & Zhao, P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Research, 39(2), 155–159.
Ni, W.-J., & Leng, X.-M. (2015). Dynamic miRNA–mRNA paradigms: New faces of miRNAs. Biochemistry and Biophysics Reports, 4, 337–341. https://doi.org/10.1016/j.bbrep.2015.10.011
Brümmer, A., & Hausser, J. (2014). MicroRNA binding sites in the coding region of mRNAs: Extending the repertoire of post-transcriptional gene regulation. BioEssays, 36(6), 617–626.
Quax, T. E., Claassens, N. J., Söll, D., & van der Oost, J. (2015). Codon bias as a means to fine-tune gene expression. Molecular Cell, 59(2), 149–161.
Chakraborty, S., Mazumder, T. H., & Uddin, A. (2018). Compositional dynamics and codon usage pattern of BRCA1 gene across nine mammalian species. Genomics. https://doi.org/10.1016/j.ygeno.2018.01.013
Wan, X.-F., Zhou, J., & Xu, D. (2006). CodonO: A new informatics method for measuring synonymous codon usage bias within and across genomes. International Journal of General Systems, 35(1), 109–125.
Uddin, A., Paul, N., & Chakraborty, S. (2019). The codon usage pattern of genes involved in ovarian cancer. Annals of the New York Academy of Sciences, 1440(1), 67–78. https://doi.org/10.1111/nyas.14019
Jenkins, G. M., & Holmes, E. C. (2003). The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Research, 92(1), 1–7.
Wu, Y., Zhao, D., & Tao, J. (2015). Analysis of codon usage patterns in herbaceous peony (Paeonia lactiflora Pall.) based on transcriptome data. Genes, 6(4), 1125–1139.
Chen, K., & Rajewsky, N. (2006). Natural selection on human microRNA binding sites inferred from SNP data. Nature genetics, 38(12), 1452–1456.
Simkin, A., Geissler, R., McIntyre, A. B. R., & Grimson, A. (2020). Evolutionary dynamics of microRNA target sites across vertebrate evolution. PLOS Genetics, 16(2), e1008285. https://doi.org/10.1371/journal.pgen.1008285
Fang, Z., & Rajewsky, N. (2011). The impact of miRNA target sites in coding sequences and in 3′ UTRs. PLoS ONE, 6(3), e18067.
Lewis, B. P., Burge, C. B., & Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120(1), 15–20.
Mullany, L. E., Herrick, J. S., Wolff, R. K., & Slattery, M. L. (2016). MicroRNA Seed region length impact on target messenger RNA expression and survival in colorectal cancer. PLoS ONE, 11(4), e0154177. https://doi.org/10.1371/journal.pone.0154177
Chakraborty, S., Paul, S., Nath, D., Choudhury, Y., Ahn, Y., Cho, Y. S., & Uddin, A. (2020). Synonymous codon usage and context analysis of genes associated with pancreatic cancer: Running title: Codon usage of pancreatic cancer genes. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis, 821, 111719.
Newman, Z. R., Young, J. M., Ingolia, N. T., & Barton, G. M. (2016). Differences in codon bias and GC content contribute to the balanced expression of TLR7 and TLR9. Proceedings of the National Academy of Sciences, 113(10), E1362–E1371.
Guo, X., Gui, Y., Wang, Y., Zhu, Q.-H., Helliwell, C., & Fan, L. (2008). Selection and mutation on microRNA target sequences during rice evolution. BMC Genomics, 9(1), 454.
Chen, H., Zhang, Z., & Feng, D. (2019). Prediction and interpretation of miRNA-disease associations based on miRNA target genes using canonical correlation analysis. BMC Bioinformatics, 20(1), 404.
O’Day, E., & Lal, A. (2010). MicroRNAs and their target gene networks in breast cancer. Breast Cancer Research, 12(2), 201. https://doi.org/10.1186/bcr2484
Acknowledgements
We are thankful to Assam University, Silchar, Assam, India for providing the necessary facilities in carrying out this research work. Further, the first author (SC) acknowledges the financial support of ICMR (5/13/42/2018/NCD-II), Government of India for partly supporting this research work.
Funding
The research work is partly funded by ICMR (5/13/42/2018/NCD-II), Government of India and partly by Assam University, Silchar, Assam.
Author information
Authors and Affiliations
Contributions
SC contributed to data curation, conceptualization, project administration, formal analysis, software, visualization, supervision, methodology, and writing reviewing, and editing of the manuscript. DN contributed to data curation, writing of the original draft, analysis of data, and interpretation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no potential conflicts of interest in this study.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12033_2022_491_MOESM1_ESM.docx
Accession numbers of the overexpressed lung cancer genes (coding sequences) targeted by microRNAs: >NM_002458- MUC5B, >NM_024690- MUC16, >NM_000088- COL1A1, >NM_001278074- COL5A1, >NM_001408- CELSR2, >NM_000094- COL7A1, and >NM_001447- FAT2. Supplementary file1 (DOCX 12 kb)
Rights and permissions
About this article
Cite this article
Chakraborty, S., Nath, D. A Study on microRNAs Targeting the Genes Overexpressed in Lung Cancer and their Codon Usage Patterns. Mol Biotechnol 64, 1095–1119 (2022). https://doi.org/10.1007/s12033-022-00491-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00491-3