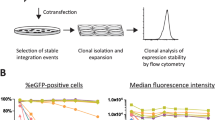
Abstract
Gene delivery to esophageal tissue could provide novel treatments for diseases, such as cancer. The Sleeping Beauty (SB) transposon system, as a natural and non-viral tool, is efficient at transferring transgene into the human genome for human cell genetic engineering. The plasmid-based SB transposon can insert into chromosomes through an accurate recombinase-mediated mechanism, providing long-term expression of transgene integrated into the target cells. In this study, we aimed to investigate the activity of ED-L2 tissue-specific promoter that was engineered from the Epstein-Barr Virus (EBV) and combined with the hyperactive SB100X transposase to achieve the stable expression of T2-Onc3 transposon in esophageal squamous epithelial cells. Here we constructed an SB transposon-based plasmid system to obtain the stable expression of transposon upon introduction of a hyperactive SB transposase under the control of tissue-specific ED-L2 promoter via the lipid-based delivery method in the cultured esophageal squamous cell carcinoma cells. Among established human and mouse cell lines, the (ED-L2)-SB100X transposase was active only in human esophageal stratified squamous epithelial and differentiated keratinocytes derived from skin (KYSE-30 and HaCaT cell lines), where it revealed high promoter activity. Data offered that the 782 bp sequence of ED-L2 promoter has a key role in its activity in vitro. The (ED-L2)-SB100X transposase mediated stable integration of T2-Onc3 in KYSE-30 cells, thereby providing further evidence of the tissue specificity of ED-L2 promoter. The KYSE-30 cells modified with the SB system integrate on average 187 copies of the T2-Onc3 transposon in its genome. In aggregate, the (ED-L2)-SB100X transposase can be efficiently applied for the tissue-specific stable expression of a transgene in human KYSE-30 cells using SB transposon.




Similar content being viewed by others
Data Availability
Data will be available on request.
Abbreviations
- SB:
-
Sleeping Beauty
- iPSC:
-
Induced pluripotent stem cell
- GEM:
-
Genetically engineered model
- PB:
-
PiggyBac
- IRs:
-
Inverted repeats
- TSP:
-
Tissue-specific promoter
- EBV:
-
Epstein-Barr virus
- PMA:
-
Phorbol 12-myristate 13-acetate
- gDNA:
-
Genomic DNA
- Ct:
-
Cycle threshold
- ALB:
-
Albumin
- MFI:
-
Mean florescence intensity
- CAG:
-
CMV enhancer/chicken beta-actin promoter
- TF:
-
Transcription factors
References
Querques, I., et al. (2019). A highly soluble Sleeping Beauty transposase improves control of gene insertion. Nature biotechnology, 37(12), 1502–1512.
Vigdal, T. J., et al. (2002). Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. Journal of molecular biology, 323(3), 441–452.
Amberger, M., & Ivics, Z. (2020). Latest advances for the sleeping beauty transposon system: 23 years of insomnia but prettier than ever: Refinement and recent innovations of the sleeping beauty transposon system enabling novel, nonviral genetic engineering applications. BioEssays, 42(11), 2000136.
Izsvák, Z., & Ivics, Z. (2004). Sleeping beauty transposition: Biology and applications for molecular therapy. Molecular Therapy, 9(2), 147–156.
Katter, K., et al. (2013). Transposon-mediated transgenesis, transgenic rescue, and tissue-specific gene expression in rodents and rabbits. The FASEB Journal, 27(3), 930–941.
Hudecek, M., et al. (2017). Going non-viral: The Sleeping Beauty transposon system breaks on through to the clinical side. Critical Reviews in Biochemistry and Molecular Biology, 52(4), 355–380.
Ivics, Z., et al. (2007). Targeted Sleeping Beauty transposition in human cells. Molecular therapy, 15(6), 1137–1144.
Glover, D. J., Lipps, H. J., & Jans, D. A. (2005). Towards safe, non-viral therapeutic gene expression in humans. Nature Reviews Genetics, 6(4), 299–310.
Zayed, H., et al. (2004). Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Molecular Therapy, 9(2), 292–304.
Wilber, A., et al. (2007). Messenger RNA as a source of transposase for sleeping beauty transposon–mediated correction of hereditary tyrosinemia type I. Molecular Therapy, 15(7), 1280–1287.
Jin, Z., et al. (2011). The hyperactive Sleeping Beauty transposase SB100X improves the genetic modification of T cells to express a chimeric antigen receptor. Gene therapy, 18(9), 849–856.
Furushima, K., et al. (2012). Insertional mutagenesis by a hybrid piggyBac and sleeping beauty transposon in the rat. Genetics, 192(4), 1235–1248.
Cocchiarella, F., et al. (2016). Transcriptionally regulated and nontoxic delivery of the hyperactive Sleeping Beauty transposase. Molecular Therapy-Methods & Clinical Development, 3, 16038.
Izsvák, Z., Ivics, Z., & Plasterk, R. H. (2000). Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. Journal of Molecular Biology, 302(1), 93–102.
Staunstrup, N. H., et al. (2009). Hybrid lentivirus-transposon vectors with a random integration profile in human cells. Molecular Therapy, 17(7), 1205–1214.
Wang, K., Kievit, F. M., & Zhang, M. (2016). Nanoparticles for cancer gene therapy: Recent advances, challenges, and strategies. Pharmacological Research, 114, 56–66.
Chen, C., et al. (2018). Promoter-operating targeted expression of gene therapy in cancer: Current stage and prospect. Molecular Therapy-Nucleic Acids, 11, 508–514.
Okabe, S. (1999). Gene expression in transgenic mice using neural promoters. Current Protocols in Neuroscience. https://doi.org/10.1002/0471142301.ns0316s07
Miskey, C., et al. (2005). DNA transposons in vertebrate functional genomics. Cellular and Molecular Life Sciences, 62(6), 629–641.
Nakagawa, H., Inomoto, T., & Rustgi, A. K. (1997). A CACCC box-like cis-regulatory element of the Epstein-Barr virus ED-L2 promoter interacts with a novel transcriptional factor in tissue-specific squamous epithelia. Journal of Biological Chemistry, 272(26), 16688–16699.
Mahmoudian, R. A., Farshchian, M., & Abbaszadegan, M. R. (2021). Genetically engineered mouse models of esophageal cancer. Experimental Cell Research, 406(2), 112757.
Nakagawa, H., et al. (1997). The targeting of the cyclin D1 oncogene by an Epstein-Barr virus promoter in transgenic mice causes dysplasia in the tongue, esophagus and forestomach. Oncogene, 14(10), 1185–1190.
Mueller, A., et al. (1997). A transgenic mouse model with cyclin D1 overexpression results in cell cycle, epidermal growth factor receptor, and p53 abnormalities. Cancer Research, 57(24), 5542–5549.
Opitz, O. G., et al. (2002). A mouse model of human oral-esophageal cancer. The Journal of Clinical Investigation, 110(6), 761–769.
Stairs, D. B., et al. (2011). Deletion of p120-catenin results in a tumor microenvironment with inflammation and cancer that establishes it as a tumor suppressor gene. Cancer Cell, 19(4), 470–483.
Kikuchi, K., et al. (2017). Epstein-Barr virus (EBV)-associated epithelial and non-epithelial lesions of the oral cavity. Japanese Dental Science Review, 53(3), 95–109.
Dupuy, A. J., et al. (2009). A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Research, 69(20), 8150–8156.
Kim, J. H., et al. (2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE, 6(4), e18556.
Khaleghizadeh, M., et al. (2018). Ectopic expression of human DPPA2 gene in ESCC cell line using retroviral system. Avicenna Journal of Medical Biotechnology, 10(2), 75.
Ayyoob, K., et al. (2016). Authentication of newly established human esophageal squamous cell carcinoma cell line (YM-1) using short tandem repeat (STR) profiling method. Tumor Biology, 37(3), 3197–3204.
Mahmoudian, R. A., Farshchian, M., & Abbaszadegan, M. R. (2020). Evaluation and optimization of lipofectamine 3000 reagents for transient gene expression in KYSE-30 esophagus cancer cell line. Archives of Medical Laboratory Sciences, 6, 1–9.
Mahmoudian, R. A., et al. (2021). Interaction between LINC-ROR and stemness state in gastric cancer cells with helicobacter pylori Infection. Iranian Biomedical Journal, 25(3), 157–168.
Han, X., et al. (2012). Selection of reliable reference genes for gene expression studies using real-time PCR in tung tree during seed development. PLoS ONE, 7(8), e43084.
Schmittgen, T. D., & Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C T method. Nature Protocols, 3(6), 1101.
Bustin, S. A., et al. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Oxford University Press.
Raeisossadati, R., et al. (2011). Quantitative analysis of TEM-8 and CEA tumor markers indicating free tumor cells in the peripheral blood of colorectal cancer patients. International Journal of Colorectal Disease, 26(10), 1265–1270.
Kawakami, K., Largaespada, D. A., & Ivics, Z. (2017). Transposons as tools for functional genomics in vertebrate models. Trends in Genetics, 33(11), 784–801.
Narayanavari, S. A., et al. (2017). Sleeping Beauty transposition: From biology to applications. Critical Reviews in Biochemistry and Molecular Biology, 52(1), 18–44.
Kuzmin, D., et al. (2010). Novel strong tissue specific promoter for gene expression in human germ cells. BMC Biotechnology, 10(1), 1–10.
Liew, C. G., et al. (2007). Transient and stable transgene expression in human embryonic stem cells. Stem Cells, 25(6), 1521–1528.
Tabar, M. S., et al. (2015). Evaluating electroporation and lipofectamine approaches for transient and stable transgene expressions in human fibroblasts and embryonic stem cells. Cell Journal (Yakhteh), 17(3), 438.
Jenkins, T. D., et al. (1998). Transactivation of the human keratin 4 and Epstein-Barr virus ED-L2 promoters by gut-enriched Krüppel-like factor. Journal of Biological Chemistry, 273(17), 10747–10754.
Jenkins, T. D., Nakagawa, H., & Rustgi, A. K. (1997). The keratinocyte-specific Epstein-Barr virus ED-L2 promoter is regulated by phorbol 12-myristate 13-acetate through twocis-regulatory elements containing E-box and Krüppel-like factor motifs. Journal of Biological Chemistry, 272(39), 24433–24442.
Tétreault, M.-P., et al. (2016). Esophageal expression of active IκB Kinase-β in mice up-regulates tumor necrosis factor and granulocyte-macrophage colony-stimulating factor, promoting inflammation and angiogenesis. Gastroenterology, 150(7), 1609-1619.e11.
Tetreault, M. P., et al. (2010). Klf4 overexpression activates epithelial cytokines and inflammation-mediated esophageal squamous cell cancer in mice. Gastroenterology, 139(6), 2124-2134.e9.
Andl, C. D., et al. (2003). Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. Journal of Biological Chemistry, 278(3), 1824–1830.
Belur, L. R., et al. (2003). Gene insertion and long-term expression in lung mediated by the Sleeping Beauty transposon system. Molecular Therapy, 8(3), 501–507.
Fong, L. Y., et al. (2003). Combined cyclin D1 overexpression and zinc deficiency disrupts cell cycle and accelerates mouse forestomach carcinogenesis. Cancer Research, 63(14), 4244–4252.
Galla, M., et al. (2011). Avoiding cytotoxicity of transposases by dose-controlled mRNA delivery. Nucleic Acids Research, 39(16), 7147–7160.
Izsvák, Z., et al. (2009). Efficient stable gene transfer into human cells by the Sleeping Beauty transposon vectors. Methods, 49(3), 287–297.
Dalsgaard, T., et al. (2009). Shielding of sleeping beauty DNA transposon-delivered transgene cassettes by heterologous insulators in early embryonal cells. Molecular Therapy, 17(1), 121–130.
Mátés, L., et al. (2009). Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nature Genetics, 41(6), 753–761.
Grabundzija, I., et al. (2010). Comparative analysis of transposable element vector systems in human cells. Molecular Therapy, 18(6), 1200–1209.
Lajos, M., et al. (2009). Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nature Genetics, 41, 753–761.
Acknowledgements
Authors wish to thank the personnel of Division of Human Genetics of Avicenna Research Institute and ACECR-Khorasan Razavi Branch for kindest collaboration.
Funding
The results presented in this paper were part of a PhD student thesis that was supported by Mashhad University of Medical Sciences (# 921706).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest with any person or organization regarding this research.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Ethical Approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mahmoudian, R.A., Fathi, F., Farshchian, M. et al. Construction and Quantitative Evaluation of a Tissue-Specific Sleeping Beauty by EDL2-Specific Transposase Expression in Esophageal Squamous Carcinoma Cell Line KYSE-30. Mol Biotechnol 65, 350–360 (2023). https://doi.org/10.1007/s12033-022-00490-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00490-4