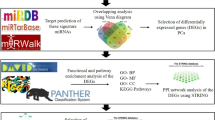
Abstract
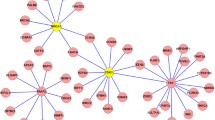
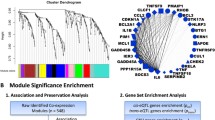
Prostate cancer (PCa) is the second most common form of cancer in men around the world. Due to its heterogeneity, presentations range from aggressive lethal disease to indolent disease. There is a need to identify core biomarkers that are important for early detection and progression, allowing a more precise method for the treatment and management of Pca. We obtained metastatic prostate cancer associated microRNA array profiles from the GSE28029 dataset in the GEO database. MicroRNA target prediction was done using the databases, TargetScanHuman, miRDB and DIANA microT, six target genes (FOXC1, CDKN1A, BIRC2, CTNND1, ELK1 and LRP8) were found to be common among the three different databases. Differential expression of the target genes was performed via the GENT2 database in the GPL96 platform (HG-U133A). Results indicated all genes were downregulated. Gene Ontology (GO) was used to perform enrichment analysis. The GO enrichment analysis indicated that the downregulated genes were enriched in cellular response to gamma radiation, regulation of transcription and response to drugs as well as protein binding and receptor signaling protein activity. The study suggested that CDKN1A, FOXC1 and BIRC2 might be core genes for prostate cancer that play an important role in its diagnosis, development and progression.
Graphic Abstract








Similar content being viewed by others
References
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., & Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68(6), 394–424. https://doi.org/10.3322/caac.21492
Ferlay, J., Lam, F., Colombet, M., Mery, L., Pineros, M., Znaor, A., Soerjomataram, I., et al. (2019). Global cancer observatory: Cancer today. Lyon: International Agency for Research on Cancer.
Stelcer, E., Konkol, M., Głȩboka, A., & Suchorska, W. M. (2019). Liquid biopsy in oligometastatic prostate cancer-A biologist’s point of view. Frontiers in Oncology, 9, 775. https://doi.org/10.3389/fonc.2019.00775
Barry, M. J. (2001). Clinical practice. Prostate-specific-antigen testing for early diagnosis of prostate cancer. The New England Journal of Medicine, 344(18), 1373–1377. https://doi.org/10.1056/NEJM200105033441806
Määttänen, L., Hakama, M., Tammela, T. L., Ruutu, M., Ala-Opas, M., Juusela, H., Martikainen, P., Stenman, U. H., & Auvinen, A. (2007). Specificity of serum prostate-specific antigen determination in the Finnish prostate cancer screening trial. British Journal of Cancer, 96(1), 56–60. https://doi.org/10.1038/sj.bjc.6603522
Basch, E., Loblaw, D. A., Oliver, T. K., Carducci, M., Chen, R. C., Frame, J. N., et al. (2014). Systemic therapy in men with metastatic castration-resistant prostate cancer: American Society of Clinical Oncology and Cancer Care Ontario clinical practice guideline. Journal of Clinical Oncology, 32(30), 3436–3448. https://doi.org/10.1200/JCO.2013.54.8404
Etzioni, R., Penson, D. F., Legler, J. M., di Tommaso, D., Boer, R., Gann, P. H., & Feuer, E. J. (2002). Overdiagnosis due to prostate-specific antigen screening: Lessons from U.S. prostate cancer incidence trends. Journal of the National Cancer Institute, 94(13), 981–990. https://doi.org/10.1093/jnci/94.13.981
Andriole, G. L., Crawford, E. D., Grubb, R. L., Buys, S. S., Chia, D., Church, T. R., et al. (2009). Mortality results from a randomized prostate-cancer screening trial. New England Journal of Medicine, 360(13), 1310–1319.
Vickers, A. J., Sjoberg, D. D., Ulmert, D., Vertosick, E., Roobol, M. J., Thompson, I., et al. (2014). Empirical estimates of prostate cancer overdiagnosis by age and prostate-specific antigen. BMC Medicine, 12(1), 1–7.
Suzuki, H., Maruyama, R., Yamamoto, E., & Kai, M. (2013). Epigenetic alteration and microRNA dysregulation in cancer. Frontiers in Genetics, 4, 258.
Mizoguchi, A., Takayama, A., Arai, T., Kawauchi, J., & Sudo, H. (2018). MicroRNA-8073: Tumor suppressor and potential therapeutic treatment. PLoS ONE, 13(12), e0209750.
Adams, B. D., Kasinski, A. L., & Slack, F. J. (2014). Aberrant regulation and function of microRNAs in cancer. Current Biology, 24, R762-776.
Ruan, L., Xie, Y., Liu, F., & Chen, X. (2018). Serum miR-1181 and miR-4314 associated with ovarian cancer: MiRNA microarray data analysis for a pilot study. European Journal of Obstetrics, Gynecology, and Reproductive Biology, 222, 31–38.
Yang, H., Wang, L., Tang, X., & Bai, W. (2017). MiR-203a suppresses cell proliferation by targeting E2F transcription factor 3 in human gastric cancer. Oncology Letters, 14(6), 7687–7690.
Chen, Z. H., Zhang, G. L., Li, H. R., Luo, J. D., Li, Z. X., Chen, G. M., et al. (2012). A panel of five circulating microRNAs as potential biomarkers for prostate cancer. Prostate, 72(13), 1443–1452.
Kojima, S., Goto, Y., & Naya, Y. (2017). The roles of microRNAs in the progression of castration-resistant prostate cancer. Journal of Human Genetics, 62, 25–31.
Kanwal, R., Plaga, A. R., Liu, X., Shukla, G. C., & Gupta, S. (2017). MicroRNAs in prostate cancer: Functional role as biomarkers. Cancer Letters, 407, 9–20.
Lyu, J., Zhao, L., Wang, F., Ji, J., Cao, Z., Xu, H., et al. (2019). Discovery and validation of serum MicroRNAs as early diagnostic biomarkers for prostate cancer in Chinese population. BioMed Research International. https://doi.org/10.1155/2019/9306803
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2013). NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Research, 41(D1), D991.
Agarwal, V., Bell, G. W., Nam, J. W., & Bartel, D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife, 4, 2015.
Wong, N., & Wang, X. (2015). miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Research, 43(D1), D146–D152.
Paraskevopoulou, M. D., Georgakilas, G., Kostoulas, N., Vlachos, I. S., Vergoulis, T., Reczko, M., et al. (2013). DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Research, 41, 1.
Park, S. J., Yoon, B. H., Kim, S. K., & Kim, S. Y. (2019). GENT2: An updated gene expression database for normal and tumor tissues. BMC Medical Genomics, 12(Suppl 5), 101–101. https://doi.org/10.1186/s12920-019-0514-7
Busk, P. K. (2014). A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinformatics, 15(1), 1–9.
Balcells, I., Cirera, S., & Busk, P. K. (2011). Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers. BMC Biotechnology, 11(1), 1–11.
Jiao, X., Sherman, B. T., Huang, D. W., Stephens, R., Baseler, M. W., Lane, H. C., et al. (2012). DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics, 28(13), 1805–1806.
Huang, Y., Zou, Q., Song, H., Song, F., Wang, L., Zhang, G., et al. (2010). A study of miRNAs targets prediction and experimental validation. Protein & Cell, 1, 979–986.
Huang, D. W., Sherman, B. T., & Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4(1), 44–57.
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2015). STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Research, 43(D1), D447–D452.
Watahiki, A., Wang, Y., Morris, J., Dennis, K., O’Dwyer, H. M., Gleave, M., et al. (2011). MicroRNAs associated with metastatic prostate cancer. PLoS ONE, 6(9), 24950.
Grün, D., Wang, Y. L., Langenberger, D., Gunsalus, K. C., & Rajewsky, N. (2005). MicroRNA target predictions across seven drosophilo species and comparison to mammalian targets. PLoS Computational Biology, 1(1), 0051–0066.
Lall, S., Grün, D., Krek, A., Chen, K., Wang, Y. L., Dewey, C. N., et al. (2006). A genome-wide map of conserved MicroRNA targets in C. elegans. Current Biology, 16(5), 460–471.
Gaidatzis, D., van Nimwegen, E., Hausser, J., & Zavolan, M. (2007). Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics, 8, 1–22.
Livak, K. J., & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔct method. Methods, 25(4), 402–408.
Nelson, W. G., De Marzo, A. M., & DeWeese, T. L. (2001). The molecular pathogenesis of prostate cancer: Implications for prostate cancer prevention. Urology, 57(4), 39–45.
Ambs, S., Prueitt, R. L., Yi, M., Hudson, R. S., Howe, T. M., Petrocca, F., et al. (2008). Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Research, 68(15), 6162–6170.
Leite, K. R. M., Tomiyama, A., Reis, S. T., Sousa-Canavez, J. M., Sañudo, A., Camara-Lopes, L. H., et al. (2013). MicroRNA expression profiles in the progression of prostate cancer-from high-grade prostate intraepithelial neoplasia to metastasis. Urologic Oncology: Seminars and Original Investigations, 31(6), 796–801.
Coleman, W. B. (2018). Molecular pathogenesis of prostate cancer. Molecular pathology: The molecular basis of human disease (pp. 555–568). Amsterdam: Elsevier.
Bradner, J. E., Hnisz, D., & Young, R. A. (2017). Transcriptional addiction in cancer. Cell, 168, 629–643. https://doi.org/10.1016/j.cell.2016.12.013
Wang, Y., Shao, C., Shi, C. H., Zhang, L., Yue, H. H., Wang, P. F., et al. (2005). Change of the cell cycle after flutamide treatment in prostate cancer cells and its molecular mechanism. Asian Journal of Andrology, 7(4), 375–380.
Devreotes, P., & Horwitz, A. R. (2015). Signaling networks that regulate cell migration. Cold Spring Harbor Perspectives in Biology, 7(8), 1.
Sever, R., & Brugge, J. S. (2015). Signal transduction in cancer. Cold Spring Harbor Perspectives in Biology, 5(4), a006098.
Sorrentino, G., Ruggeri, N., Zannini, A., Ingallina, E., Bertolio, R., Marotta, C., et al. (2017). Glucocorticoid receptor signalling activates YAP in breast cancer. Nature Communication, 8, 1–14.
Yu, S., Sun, L., Jiao, Y., & Lee, L. T. O. (2018). The role of G protein-coupled receptor kinases in cancer. International Journal of Biological Sciences., 14, 189–203.
Shah, K., Gagliano, T., Garland, L., O’Hanlon, T., Bortolotti, D., Gentili, V., et al. (2020). Androgen receptor signaling regulates the transcriptome of prostate cancer cells by modulating global alternative splicing. Oncogene, 39(39), 6172–6189. https://doi.org/10.1038/s41388-020-01429-2
Corces, M. R., Granja, J. M., Shams, S., Louie, B. H., Seoane, J. A., Zhou, W., et al. (2018). The chromatin accessibility landscape of primary human cancers. Science, 362, 6413.
Chaudhary, K. S., Abel, P. D., & Lalani, E. N. (1999). Role of the Bcl-2 gene family in prostate cancer progression and its implications for therapeutic intervention. Environmental Health Perspectives, 107(Suppl 1), 49.
Renner, W., Langsenlehner, U., Krenn-Pilko, S., Eder, P., & Langsenlehner, T. (2017). BCL2-Genotypen und Überleben bei Prostatakrebs. Strahlentherapie und Onkol, 193(6), 466–471. https://doi.org/10.1007/s00066-017-1126-9
Miller, D. R., Ingersoll, M. A., & Lin, M. F. (2018). ErbB-2 signaling in advanced prostate cancer progression and potential therapy. Endocrine-Related Cancer, 26, R195-209. https://doi.org/10.1530/ERC-19-0009
Edwards, I. J. (2012). Proteoglycans in prostate cancer. Nature Reviews Urology, 9, 196–206.
Suhovskih, A. V., Mostovich, L. A., Kunin, I. S., Boboev, M. M., Nepomnyashchikh, G. I., Aidagulova, S. V., et al. (2013). Proteoglycan expression in normal human prostate tissue and prostate cancer. ISRN Oncology, 2013, 1–9.
Warfel, N. A., & El-Deiry, W. S. (2013). P21WAF1 and tumourigenesis: 20 Years after. Current Opinion in Oncology, 25, 52–58.
Eastham, J. A., Hall, S. J., Sehgal, I., Wang, J., Timme, T. L., Yang, G., Connell-Crowley, L., Elledge, S. J., Zhang, W. W., Harper, J. W., et al. (1995). In vivo gene therapy with p53 or p21 adenovirus for prostate cancer. Cancer Research, 55(22), 5151–5155.
Li, S., Wang, C., Yu, X., Wu, H., Hu, J., Wang, S., et al. (2017). MiR-3619-5p inhibits prostate cancer cell growth by activating CDKN1A expression. Oncology Reports, 37(1), 241–248.
Yang, Z., Jiang, S., Cheng, Y., Li, T., Hu, W., Ma, Z., Chen, F., & Yang, Y. (2017). FOXC1 in cancer development and therapy: Deciphering its emerging and divergent roles. Therapeutic Advances in Medical Oncology, 9(12), 797–816. https://doi.org/10.1177/1758834017742576
Elian, F. A., Yan, E., & Walter, M. A. (2017). FOXC1, the new player in the cancer sandbox. Oncotarget, 9(8), 8165–8178. https://doi.org/10.18632/oncotarget.22742
van der Heul-Nieuwenhuijsen, L., Dits, N. F., & Jenster, G. (2009). Gene expression of forkhead transcription factors in the normal and diseased human prostate. BJU International, 103(11), 1574–1580. https://doi.org/10.1111/j.1464-410X.2009.08351.x
Parajuli, K. R., Zhang, Q., Liu, S., Patel, N. K., Lu, H., Zeng, S. X., et al. (2014). Methoxyacetic acid suppresses prostate cancer cell growth by inducing growth arrest and apoptosis. American Journal of Clinical and Experimental Urology, 2(4), 300–312.
Luk, S. U., Xue, H., Cheng, H., Lin, D., Gout, P. W., Fazli, L., Collins, C. C., Gleave, M. E., & Wang, Y. (2014). The BIRC6 gene as a novel target for therapy of prostate cancer: Dual targeting of inhibitors of apoptosis. Oncotarget, 5(16), 6896–6908. https://doi.org/10.18632/oncotarget.2229
Acknowledgements
The authors thank the Biotechnology Department at UWC, the NIC and DST Minetk for their aid in the project.
Funding
Funding for part of this work was provided for by the Nanotechnology Innovation (NIC) Centre with the aid of the Department of Science and Technology (DST) Mintek South Africa.
Author information
Authors and Affiliations
Contributions
The corresponding author conceived the idea, performed the experiments, guided the aim of study, interpreted the project results and drafted the manuscript. The other authors supervised the project and reviewed the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lombe, C.P., Meyer, M. & Pretorius, A. Bioinformatics Prediction and Analysis of MicroRNAs and Their Targets as Biomarkers for Prostate Cancer: A Preliminary Study. Mol Biotechnol 64, 401–412 (2022). https://doi.org/10.1007/s12033-021-00414-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-021-00414-8