Abstract
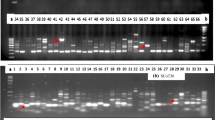
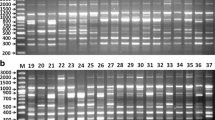
Studies on genetic diversity could enhance taxonomic authentication and evolutionary relationship among the species of Ocimum. Therefore, diversity among 36 Ocimum accessions representing species from different regions of world were analyzed using Start Codon-Targeted Polymorphism (SCoT) and inter-simple sequences repeat (ISSR) marker. Marker systems used in this study was potentially targeted the different regions of the genome and included 18 SCoT and 15 ISSR primers, which showed successful amplification profile for Ocimum. Between these two, SCoT revealed the highest mean value of percentage of Polymorphism (84.6%), polymorphic information content (PIC, 0.65), and resolving power (Rp, 8.80), which were higher than ISSR. A total of 140 and 111 amplicons were obtained with SCoT and ISSR marker. The Mantel test indicted a significant correlation (r2 = 0.44) between ISSR and SCoT, which suggested a common genetical background among the accessions. The principal coordinate study showed the selection of different Ocimum genotypes by the cluster analysis. This study will help and support identification, genetic mapping, and molecular ecology to enhance the breeding program's efficiency for developing elite varieties to meet industrial demand globally. The present study is the first report of the genetic diversity, and relationship determination with SCoT-based molecular marker among Ocimum accessions.









Similar content being viewed by others
References
Prakash, P., & Gupta, N. (2005). Therapeutic uses of Ocimum sanctum Linn (Tulsi) with a note on eugenol and its pharmacological actions: A short review. Indian Journal of Physiology and Pharmacology, 49(2), 125–131.
Chowdhury, T., Mandal, A., Roy, S. C., & De Sarker, D. (2017). Diversity of the genus Ocimum (Lamiaceae) through morpho-molecular (RAPD) and chemical (GC–MS) analysis. Journal of Genetic Engineering and Biotechnology, 15(1), 275–286. https://doi.org/10.1016/j.jgeb.2016.12.004
Bhattacharjya, D., Adhikari, S., Biswas, A., Bhuimali, A., Ghosh, P., & Saha, S. (2019). Ocimum phytochemicals and their potential impact on human health. Phytochemicals in Human Health. https://doi.org/10.5772/intechopen.88555
Pandey, A. K., Singh, P., & Tripathi, N. N. (2014). Chemistry and bioactivities of essential oils of some Ocimum species: An overview. Asian Pacific Journal of Tropical Biomedicine, 4(9), 682–694. https://doi.org/10.12980/APJTB.4.2014C77
Aldarkazali, Rihan, Carne, & Fuller. (2019). The growth and development of Sweet Basil (Ocimum basilicum) and Bush Basil (Ocimum minimum) grown under three light regimes in a controlled environment. Agronomy, 9(11), 743. https://doi.org/10.3390/agronomy9110743
Pushpangadan, P., & George, V. (2012). 4—Basil. In K. V. Peter (Ed.), Handbook of herbs and spices (second edition) (pp. 55–72). Woodhead Publishing. https://doi.org/10.1533/9780857095671.55
Cohen, M. M. (2014). Tulsi—Ocimum sanctum: A herb for all reasons. Journal of Ayurveda and Integrative Medicine, 5(4), 251–259. https://doi.org/10.4103/0975-9476.146554
Bast, F., Rani, P., & Meena, D. (2014, January 2). Chloroplast DNA phylogeography of Holy Basil (Ocimum tenuiflorum) in Indian subcontinent. The Scientific World Journal. https://doi.org/10.1155/2014/847482
Kumar, A., Mishra, P., Baskaran, K., Shukla, A. K., Shasany, A. K., & Sundaresan, V. (2016). Higher efficiency of ISSR markers over plastid psbA-trnH region in resolving taxonomical status of genus Ocimum L. Ecology and Evolution, 6(21), 7671–7682. https://doi.org/10.1002/ece3.2483
Patel, H. K., Fougat, R. S., Kumar, S., Mistry, J. G., & Kumar, M. (2015). Detection of genetic variation in Ocimum species using RAPD and ISSR markers. 3 Biotech, 5(5), 697–707. https://doi.org/10.1007/s13205-014-0269-y
Alves, R. P., Silva, A. V. C., Almeida-Pereira, C. S., Costa, T. S., Alvares-Carvalho, S. V., Arrigoni-Blank, M. D. F., & Blank, A. F. (2019). Genetic divergence in basil cultivars and hybrids. Horticultura Brasileira, 37(2), 180–187. https://doi.org/10.1590/s0102-053620190208
Thakur, J., Dwivedi, M. D., Sourabh, P., Uniyal, P. L., & Pandey, A. K. (2016). Genetic homogeneity revealed using SCoT, ISSR and RAPD markers in micropropagated Pittosporum eriocarpum Royle—An endemic and endangered medicinal plant. PLoS ONE, 11(7), e0159050. https://doi.org/10.1371/journal.pone.0159050
Etminan, A., Pour-Aboughadareh, A., Noori, A., Ahmadi-Rad, A., Shooshtari, L., Mahdavian, Z., & Yousefiazar-Khanian, M. (2018). Genetic relationships and diversity among wild Salvia accessions revealed by ISSR and SCoT markers. Biotechnology & Biotechnological Equipment, 32(3), 610–617. https://doi.org/10.1080/13102818.2018.1447397
Khanuja, S. P. S., Shasany, A. K., Darokar, M. P., & Kumar, S. (1999). Rapid isolation of DNA from dry and fresh samples of plants producing large amounts of secondary metabolites and essential oils. Plant Molecular Biology Reporter, 17(1), 74–74. https://doi.org/10.1023/A:1007528101452
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89(3), 583–590.
Mantel, N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Research, 27(2), 209–220.
Peakall, R., & Smouse, P. E. (2006). genalex 6: Genetic analysis in excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6(1), 288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Tiwari, G., Singh, R., Singh, N., Choudhury, D. R., Paliwal, R., Kumar, A., & Gupta, V. (2016). Study of arbitrarily amplified (RAPD and ISSR) and gene targeted (SCoT and CBDP) markers for genetic diversity and population structure in Kalmegh [Andrographis paniculata (Burm. f.) Nees]. Industrial Crops and Products, 86, 1–11. https://doi.org/10.1016/j.indcrop.2016.03.031
Dhawan, S. S., Shukla, P., Gupta, P., & Lal, R. K. (2016). A cold-tolerant evergreen interspecific hybrid of Ocimum kilimandscharicum and Ocimum basilicum: analyzing trichomes and molecular variations. Protoplasma, 253(3), 845–855. https://doi.org/10.1007/s00709-015-0847-9
Dhawan, S. S., Gupta, P., Mishra, A., Singh, S. K., & Chauhan, H. S. (2018). Genetic differentiation within Vetiveria zizanioides: selection and breeding of commercial perfumery grass Vetiver. Journal of Herbs, Spices & Medicinal Plants. https://doi.org/10.1080/10496475.2018.1493015
Gupta, P., Mishra, A., Yadav, A., & Dhawan, S. S. (2018). Inter and intra-specific molecular and chemical diversity of elite accessions of aromatic grasses Cymbopogons. Journal of Applied Research on Medicinal and Aromatic Plants, 11, 54–60. https://doi.org/10.1016/j.jarmap.2018.10.005
Luo, C., He, X., Chen, H., Ou, S., Gao, M., Brown, J. S., Tondo, C. T., & Schnell, R. J. (2011). Genetic diversity of mango cultivars estimated using SCoT and ISSR markers. Biochemical Systematics and Ecology, 39(4–6), 676–684. https://doi.org/10.1016/j.bse.2011.05.023
Gogoi, B., Wann, S. B., & Saikia, S. P. (2020). Comparative assessment of ISSR, RAPD, and SCoT markers for genetic diversity in Clerodendrum species of North East India. Molecular Biology Reports, 47(10), 7365–7377. https://doi.org/10.1007/s11033-020-05792-x
Amom, T., Tikendra, L., Apana, N., Goutam, M., Sonia, P., Koijam, A. S., Potshangbam, A. M., Rahaman, H., & Nongdam, P. (2020). Efficiency of RAPD, ISSR, iPBS, SCoT and phytochemical markers in the genetic relationship study of five native and economical important bamboos of North-East India. Phytochemistry, 174, 112330. https://doi.org/10.1016/j.phytochem.2020.112330
Safari, H., Zebarjadi, A., Kahrizi, D., & Jafari, A. A. (2019). The study of inter-specific relationships of Bromus genus based on SCoT and ISSR molecular markers. Molecular Biology Reports, 46(5), 5209–5223. https://doi.org/10.1007/s11033-019-04978-2
Gao, Y., Zhu, Y.-Q., Tong, Z., Xu, Z., Jiang, X., & Huang, C. (2014). Analysis of genetic diversity and relationships among genus Lycoris based on start codon targeted (SCoT) marker. Biochemical Systematics and Ecology, 57, 221–226. https://doi.org/10.1016/j.bse.2014.08.002
Srivastava, A., Gupta, S., Shanker, K., Gupta, N., Gupta, A. K., & Lal, R. K. (2020). Genetic diversity in Indian poppy (P. somniferum L.) germplasm using multivariate and SCoT marker analyses. Industrial Crops and Products, 144, 112050. https://doi.org/10.1016/j.indcrop.2019.112050
Sugawara, Y. (2000). Odor distinctiveness between enantiomers of linalool: Difference in perception and responses elicited by sensory test and forehead surface potential wave measurement. Chemical Senses, 25(1), 77–84. https://doi.org/10.1093/chemse/25.1.77
Dhawan, S. S., Mishra, A., Gupta, P., Bahl, J. R., & Bansal, R. P. (2018). Phylogentic relationship of cold tolerant Mentha arvensis variety ‘CIM Kranti’ with some released varieties as assessed through physiological and molecular analysis. Journal of Applied Research on Medicinal and Aromatic Plants. https://doi.org/10.1016/j.jarmap.2018.06.004
Igwe, D. O., Afiukwa, C. A., Ubi, B. E., Ogbu, K. I., Ojuederie, O. B., & Ude, G. N. (2017). Assessment of genetic diversity in Vigna unguiculata L. (Walp) accessions using inter-simple sequence repeat (ISSR) and start codon targeted (SCoT) polymorphic markers. BMC Genetics, 18(1), 98. https://doi.org/10.1186/s12863-017-0567-6
Pakseresht, F., Talebi, R., & Karami, E. (2013). Comparative assessment of ISSR, DAMD and SCoT markers for evaluation of genetic diversity and conservation of landrace chickpea (Cicer arietinum L.) genotypes collected from north-west of Iran. Physiology and Molecular Biology of Plants, 19(4), 563–574. https://doi.org/10.1007/s12298-013-0181-7
Acknowledgements
We are thankful for support and facilities provided by Director, CSIR-CIMAP, Lucknow, India. PG acknowledges the Academy of Scientific and Innovative Research (AcSIR) for registration of Ph.D. program.
Funding
This work was supported by CSIR- Aroma mission HCP 0007. CIMAP Communication Number CIMAP/PUB/2020/OCT/109.
Author information
Authors and Affiliations
Contributions
PG carried out sample collection, design and execution of data integration, manuscript preparation. AM performed assisted in data and software analysis. RKL helped in field experiments. SSD was responsible for conceptualization, designing of the experiments, data analysis, data integration, drafting and finalization of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors confirm that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gupta, P., Mishra, A., Lal, R.K. et al. DNA Fingerprinting and Genetic Relationships Similarities Among the Accessions/Species of Ocimum Using SCoT and ISSR Markers System. Mol Biotechnol 63, 446–457 (2021). https://doi.org/10.1007/s12033-021-00316-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-021-00316-9