Abstract
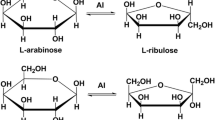
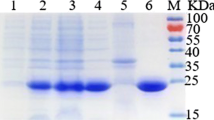
d-Tagatose is a ketohexose, which presents unique properties as a low-calorie functional sweetener possessing a sweet flavor profile similar to d-sucrose and having no aftertaste. Considered a generally recognized as safe (GRAS) substance by FAO/WHO, d-tagatose can be used as an intermediate for the synthesis of other optically active compounds as well as an additive in detergent, cosmetic, and pharmaceutical formulations. This study reports important features for l-arabinose isomerase (EC 5.3.1.4) (L-AI) use in industry. We describe arabinose (araA) gene virulence analysis, gene isolation, sequencing, cloning, and heterologous overexpression of L-AI from the food-grade GRAS bacterium Enterococcus faecium DBFIQ E36 in Escherichia coli and assess biochemical properties of this recombinant enzyme. Recombinant L-AI (rL-AI) was one-step purified to homogeneity by Ni2+-agarose resin affinity chromatography and biochemical characterization revealed low identity with both thermophilic and mesophilic L-AIs but high degree of conservation in residues involved in substrate recognition. Optimal conditions for rL-AI activity were 50 °C, pH 5.5, and 0.3 mM Mn2+, exhibiting a low cofactor concentration requirement and an acidic optimum pH. Half-life at 45 °C and 50 °C were 1427 h and 11 h, respectively, and 21.5 h and 39.5 h at pH 4.5 and 5.6, respectively, showing the high stability of the enzyme in the presence of a metallic cofactor. Bioconversion yield for d-tagatose biosynthesis was 45% at 50 °C after 48 h. These properties highlight the technological potential of E. faecium rL-AI as biocatalyst for d-tagatose production.






Similar content being viewed by others
References
Troyano, E., Villamiel, M., Olano, A., Sanz, J., & Martínez-Castro, I. (1996). Monosaccharides and Myo-inositol in commercial milks. Journal of Agricultural and Food Chemistry, 44, 815–817.
Mendoza, M. R., Olano, A., & Villamiel, M. (2005). Chemical indicators of heat treatment in fortified and special milks. Journal of Agricultural and Food Chemistry, 53, 2995–2999.
Bertelsen, H., Jensen, B. B., Buemann, B.. d-Tagatose (1999). A novel low-caloric bulk sweetener with prebiotic properties. World Review of Nutrition and Dietetics, 85, 98–109.
Patra, F., Tomar, S. K., & Arora, S. (2009). Technological and functional applications of low-calorie sweeteners from lactic acid bacteria. Journal of Food Science, 74, 16–23.
Bertelsen, H., Andersen, H., & Tvede, M. (2001). Fermentation of d-tagatose by human intestinal bacteria and dairy lactic acid bacteria. Microbial Ecology in Health and Disease, 3, 87–95.
Mortensen, P. B. 1999. In vitro fermentation of tagatose to short-chain fatty acids (SCFA) and D + l-lactate in human faecal homogenates, internal report.
Moore, M. C. (2006). Drug evaluation: Tagatose in the treatment of Type 2 diabetes and obesity. Current Opinion in Investigational Drugs, 7, 924–935.
Livesey, G. (2003). Health potential of polyols as sugar replacers, with emphasis on low glycaemic properties. Nutrition Research Reviews, 16, 163–191.
Lu, Y., Levin, G. V., & Donner, T. W. (2008). Tagatose, a new antidiabetic and obesity control drug. Diabetes, Obesity and Metabolism, 10, 109–134.
Levin, G. V. (2002). Tagatose, the new GRAS sweetener and health product. Journal of Medicinal Food, 5, 23–36.
Wong, D. (2000). Sweetener determined safe in drugs, mouthwashes, and toothpastes. Dentistry Today, 19, 34–35.
Bär, A. (2004). d-tagatose, Dossier prepared and submitted on behalf of Arla food ingredients, Amba, Viby, Denmark, for evaluation pursuant to EU novel foods regulation (EC) 258/97 by the UK advisory committee on novel foods and processes, bioresco, food scientific and regulatory services.
Favara, A. F. & Federal Drug Administration (2003). Food labelling: Health claims; d-tagatose and dental caries. Final rule. Federal Register, 68, 39831–39833.
Beadle, J. R., Saunder, J. P., & Wajada, T. J. (1992). Process for manufacturing tagatose. US Patent 5.078.796.
Freimund, S., Huwig, A., Giffhorn, F., & Köpper, S. (1996). Convenient chemo-enzymatic synthesis of d-tagatose. Journal of Carbohydrate Chemistry, 15, 115–120.
Yun, M., Moon, H. R., Kim, H. O., Choi, W. J., Kim, Y. C., Park, C. S., & Jeong, L. S. (2005). A highly efficient synthesis of unnatural l-sugars from d-ribose. Tetrahedron Letters, 46, 5903–5905.
Kim, H. J., Kang, S. Y., Park, J. J., & Kim, P. (2011). Novel activity of UDP-galactose-4-epimerase for free monosaccharide and activity improvement by active site-saturation mutagenesis. Applied Biochemistry and Biotechnology, 163, 444–451.
Oh, D. K. (2007). Tagatose: Properties, applications, and biotechnological processes. Applied Microbiology and Biotechnology, 76, 1–8.
Yeom, S. J., Kim, N. H., Park, C. S., & Oh, D. K. (2009). l-ribose production from l-arabinose by using purified l-arabinose Isomerase and mannose-6-phosphate isomerase from Geobacillus thermodenitrificans. Applied and Environmental Microbiology, 75, 6941–6943.
Hugenholtz, J., & Smid, E. J. (2002). Nutraceutical production with food-grade microorganisms. Current Opinion in Biotechnology, 13, 497–507.
Kim, H. J., Kim, J. H., Oh, H. J., & Oh, D. K. (2006). Characterization of a mutated Geobacillus stearothermophilus l-arabinose isomerase that increases the production rate of d-tagatose. Journal of Applied Microbiology, 101, 213–221.
Yoshida, H., Yamada, M., Nishitani, T., Takada, G., Izumori, K., & Kamitori, S. (2007). Purification, crystallization and preliminary X-ray diffraction studies of d-tagatose 3-epimerase from Pseudomonas cichorii. Acta Crystallographica Section F, 63, 123–125.
Boudebbouze, S., Maguin, E., & Rhimi, M. (2011). Bacterial l-arabinose isomerases: Industrial application for d-tagatose production. Recent Patents on DNA and Gene Sequences, 5, 194–201.
Kim, P. (2004). Current studies on biological tagatose production using l-arabinose isomerase: A review and future perspective. Applied Microbiology and Biotechnology, 65, 243–249.
Cheetham, P. S. J., & Wootton, A. N. (1993). Bioconversion of d-galactose into d-tagatose. Enzyme and Microbial Technology, 15, 105–108.
Xu, Z., Li, S., Feng, X., Liang, J., & Xu, H. (2014). l-arabinose isomerase and its use for biotechnological production of rare sugars. Applied Microbiology and Biotechnology, 98, 8869–8878.
Kim, B. C., Lee, Y. H., Lee, H. S., Lee, D. W., Choe, E. A., & Pyun, Y. R. (2002). Cloning, expression and characterization of l-arabinose isomerase from Thermotoga neapolitana: Bioconversion of d-galactose to d-tagatose using the enzyme. FEMS Microbiology Letters, 212, 121–126.
Lee, D. W., Choe, E. A., Kim, S. B., Eom, S. H., Hong, Y. H., Lee, S. J., Lee, H. S., Lee, D. Y., & Pyun, Y. R. (2005). Distinct metal dependence for catalytic and structural functions in the l-arabinose isomerase from the mesophilic Bacillus halodurans and the thermophilic Geobacillus stearothermophilus. Archives of Biochemistry and Biophysics, 434, 333–343.
Rhimi, M., Ilhammami, R., Bajic, G., Boudebbouze, S., Maguin, E., Haser, R., & Aghajari, N. (2010). The acid tolerant l-arabinose isomerase from the food grade Lactobacillus sakei 23K is an attractive d-tagatose producer. Bioresource Technology, 101, 9171–9177.
Torres, P. R., Manzo, R. M., Rubiolo, A. C., Batista-Viera, F. D., & Mammarella, E. J. (2014). Purification of an l-arabinose isomerase from Enterococcus faecium DBFIQ E36 employing a biospecific affinity strategy. Journal of Molecular Catalysis B: Enzymatic, 102, 99–105.
de Sousa, M., Manzo, R. M., García, J. L., Mammarella, E. J., Gonçalves, L. R. B., & Pessela, B. C. J. (2017). Engineering the l-arabinose isomerase from Enterococcus faecium for d-tagatose synthesis. Molecules, 22, 2164.
Manzo, R. M., Sousa, M., Fenoglio, C. L., Gonçalves, L. R. B., & Mammarella, E. J. (2015). Chemical improvement of chitosan-modified beads for the immobilization of Enterococcus faecium DBFIQ E36 l-arabinose isomerase through multipoint covalent attachment approach. Journal of Industrial Microbiology and Biotechnology, 42, 1325–1340.
Manzo, R. M., Simonetta, A. C., Rubiolo, A. C., & Mammarella, E. J. (2013). Screening and selection of wild strains for l-arabinose isomerase production. Brazilian Journal of Chemical Engineering, 30, 711–720.
Warner, S. A. J. (1996). Genomic DNA isolation and lambda library construction. In: G.D. Foster and D. Twell (ed) Plant gene isolation (pp. 56–58) Hoboken: Wiley.
Foulquié Moreno, M. R., Callewaert, R., Devreese, B., Van Beeumen, J., & De Vuyst, L. (2003). Isolation and biochemical characterisation of enterocins produced by enterococci from different sources. Journal of Applied Microbiology, 94, 214–229.
Manzo, R. M., Cardoso, M. D. L. M., Tonarelli, G. G., & Simonetta, A. C. (2016). Purification of two bacteriocins produced by Enterococcus faecalis DBFIQ E24 strain isolated from raw bovine milk. International Journal of Dairy Technology, 69, 282–293.
Abouelnaga, M., Lamas, A., Quintela-Baluja, M., Osman, M., Miranda, J. M., Cepeda, A., & Franco, C. M. (2016). Evaluation of the extent of spreading of virulence factors and antibiotic resistance in Enterococci isolated from fermented and unfermented foods. Annals of Microbiology, 66, 577–585.
Lam, M. M. C., Seemann, T., Bulach, D. M., Gladman, S. L., Chen, H., Haring, V., Moore, R. J., Ballard, S., Grayson, M. L., Johnson, P. D. R., Howden, B. P., & Stinear, T. P. (2012). Comparative analysis of the first complete Enterococcus faecium genome. Journal of Bacteriology, 194, 2334–2341.
Qin, X., Galloway-Peña, J. R., Sillanpaa, J., Roh, J. H., Nallapareddy, S. R., Chowdhury, S., Bourgogne, A., Choudhury, T., Muzny, D. M., Buhay, C. J., Ding, Y., Dugan-Rocha, S., Liu, W., Kovar, C., Sodergren, E., Highlander, S., Petrosino, J. F., Worley, K. C., Gibbs, R. A., Weinstock, G. M., & Murray, B. E. (2012). Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiology, 12, 135–155.
Ewing, B., & Green, P. (1998). Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Research, 8, 186–194.
Ewing, B., Miller, L., Wendl, M. C., & Green, P. (1998). Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Research, 8, 175–185.
Gordon, D., Abaijan, C., & Green, P. (1998). Consed: A graphical tool for sequence finishing. Genome Research, 8, 195–202.
Lessard, J. C. (2013). Transformation of E. coli via electroporation. Methods in Enzymology, 529, 321–327.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Walker, J. M. (2002). The bicinchoninic acid (BCA) assay for protein quantitation. In J. M. Walker (Ed.), The protein protocols handbook (2nd edn., pp. 11–14). Totowa: Humana Press Inc.
Laemmli, U. K. (1970). Cleavage of structural protein during the assembly of the heat of bacteriophage T4. Nature, 227, 680–685.
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., & Higgins, D. G. (2007). Clustal W and clustal X version 2.0. Bioinformatics, 23, 2947–2948.
Schwede, T., Kopp, J., Guex, N., & Peitsch, M. C. (2003). SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Research, 31, 3381–3385.
Chouayekh, H., Bejar, W., Rhimi, M., Jelleli, K., Mseddi, M., & Bejar, S. (2007). Characterization of an l-arabinose isomerase from the Lactobacillus plantarum NC8 strain showing pronounced stability at acidic pH. FEMS Microbiology Letters, 277, 260–267.
Dische, Z., & Borenfreund, E. (1951). A New spectrophotometric method for the detection and determination of keto sugars and trioses. Journal of Biological Chemistry, 192, 583–587.
Salonen, N., Nyyssölä, A., Salonen, K., & Turunen, O. (2012). Bifidobacterium longum l-arabinose isomerase overexpression in Lactococcus lactis, purification, and characterization. Applied Biochemistry and Biotechnology, 168, 392–405.
Rhimi, M., Aghajari, N., Juy, M., Chouayekh, H., Maguin, E., Haser, R., & Bejar, S. (2009). Rational design of Bacillus stearothermophilus US100 l-arabinose Isomerase: Potential applications for d-tagatose production. Biochimie, 91, 650–653.
Wanarska, M., & Kur, J. (2012). A method for the production of d-tagatose using a recombinant Pichia pastoris strain secreting ß-d-GALACTOSIDASE from Arthrobacter chlorophenolicus and a recombinant l-arabinose isomerase from Arthrobacter sp. 22c. Microbial Cell Factories, 11, 113–128.
Kim, H. J., & Oh, D. K. (2005). Purification and characterization of an l-arabinose isomerase from an isolated strain of Geobacillus thermodenitrificans producing d-tagatose. Journal of Biotechnology, 120, 162–173.
Torres, P., & Batista-Viera, F. (2012). Immobilization of β-galactosidase from Bacillus circulans onto Epoxy-Activated Acrylic Supports. Journal of Molecular Catalysis B: Enzymatic, 83, 57–64.
Torres, P., & Batista-Viera, F. (2012). Improved biocatalysts based on Bacillus circulans β-galactosidase immobilized onto epoxy-activated acrylic supports: Applications in whey processing. Journal of Molecular Catalysis B: Enzymatic, 74, 230–235.
Xu, Z., Qing, Y., Li, S., Feng, X., Xu, H., & Ouyang, P. (2011). A Novel l-arabinose isomerase from Lactobacillus fermentum CGMCC2921 for d-tagatose production: Gene cloning, purification and characterization. Journal of Molecular Catalysis B: Enzymatic, 70, 1–7.
Manjasetty, B. A., & Chance, M. R. (2006). Crystal structure of Escherichia coli l-arabinose isomerase (ECAI), the putative target of biological tagatose production. Journal of Molecular Biology, 360, 297–309.
Sabia, C., De Niederhäusern, S., Guerrieri, E., Messi, P., Anacarso, I., Manicardi, G., & Bondi, M. (2008). Detection of bacteriocin production and virulence traits in vancomycin-resistant enterococci of different sources. Journal of Applied Microbiology, 104, 970–979.
Semedo, T., Santos, M. A., Lopes, M. F., Marques, J. J. F., Crespo, M. T., & Tenreiro, R. (2003). Virulence factors in food, clinical and reference enterococci: A common trait in the genus? Systematic and Applied Microbiology, 26, 13–22.
Martin, B., Garriga, M., Hugas, M., & Aymerich, T. (2005). Genetic diversity and safety aspects of enterococci from slightly fermented sausages. Journal of Applied Microbiology, 98, 1177–1190.
Men, Y., Zhu, Y., Zhang, L., Kang, Z., Izumori, K., Sun, Y., & Ma, Y. (2014). Enzymatic conversion of d-galactose to d-tagatose: Cloning, overexpression and characterization of l-arabinose isomerase from Pediococcus pentosaceus PC-5. Microbiological Research, 169, 171–178.
Fan, C., Liu, K., Zhang, T., Zhou, L., Xue, D., Jiang, B., & Mu, W. (2014). Biochemical characterization of a thermostable l-arabinose isomerase from a thermoacidophilic bacterium, Alicyclobacillus hesperidum URH17-3-68. Journal of Molecular Catalysis B: Enzymatic, 102, 120–125.
Zhang, Y., Fan, Y., Hu, H., Yang, H., Luo, X., Li, Z., Zhou, H., Ma, W., Song, Y., & Zhang, T. (2017). d-Tagatose production by Lactococcus lactis NZ9000 Cells Harboring Lactobacillus plantarum l-arabinose isomerase. Indian Journal of Pharmaceutical Education and Research, 51, 288–294.
Torres, P., & Batista-Viera, F. (2017). Immobilized trienzymatic system with enhanced stabilization for the biotransformation of lactose. Molecules, 22, 284–298.
Cheng, L., Mu, W., Zhang, T., & Jiang, B. (2010). An l-arabinose isomerase from Acidothermus cellulolyticus ATCC 43068: Cloning, expression, purification, and characterization. Applied Microbiology and Biotechnology, 86, 1089–1097.
Lee, S. J., Lee, D. W., Choe, E. A., Hong, Y. H., Kim, S. B., Kim, B. C., & Pyun, Y. R. (2005). Characterization of a thermoacidophilic l-arabinose isomerase from Alicyclobacillus acidocaldarius: role of Lys-269 in pH optimum. Applied and Environmental Microbiology, 71, 7888–7896.
Choi, J. M., Lee, Y. J., Cao, T. P., Shin, S. M., Park, M. K., Lee, H. S., di Luccio, E., Kim, S. B., Lee, S. J., Lee, S. J., Lee, S. H., & Lee, D. W. (2016). Structure of the thermophilic l-arabinose isomerase from Geobacillus kaustophilus reveals metal-mediated intersubunit interactions for activity and thermostability. Archives of Biochemistry and Biophysics, 596, 51–62.
Lee, D. W., Jang, H. J., Choe, E. A., Kim, B. C., Lee, S. J., Kim, S. B., Hong, Y. H., & Pyun, Y. R. (2004). Characterization of a thermostable l-arabinose (d-galactose) isomerase from the hyperthermophilic eubacterium Thermotoga maritima. Applied and Environmental Microbiology, 70, 1397–1404.
Patrick, J. W., & Lee, N. (1968). Purification and properties of an l-arabinose isomerase from Escherichia coli. Journal of Biological Chemistry, 243, 4312–4318.
Rhimi, M., & Bejar, S. (2006). Cloning, purification and biochemical characterization of metallic-ions independent and thermoactive l-arabinose isomerase from the Bacillus stearothermophilus US100 strain. Biochimica et Biophysica Acta, 1760, 191–199.
Staudigl, P., Haltrich, D., & Peterbauer, K. (2014). l-arabinose isomerase and d-xylose isomerase from Lactobacillus reuteri: Characterization, coexpression in the food grade host Lactobacillus plantarum, and application in the conversion od d-galactose and d-glucose. Journal of Agricultural and Food Chemistry, 62, 1617–1624.
Cheng, L., Mu, W., & Jiang, B. (2010). Thermostable l-arabinose isomerase from Bacillus stearothermophilus IAM 11001 for d-tagatose production: Gene cloning, purification and characterization. Journal of the Science of Food and Agriculture, 90, 1327–1333.
Patel, M. J., Akhani, R. C., Patel, A. T., Dedania, S. R., & Patel, D. H. (2017). A single and two step isomerization process for d-tagatose and l-ribose bioproduction using l-arabinose isomerase and d-lyxose isomerase. Enzyme and Microbial Technology, 97, 27–33.
Yamanaka, K. (1975). l-Arabinose isomerase from Lactobacillus gayonii. Methods in Enzymology, 41, 458–461.
Yoon, S. H., Kim, P., & Oh, D. K. (2003). Properties of l-arabinose isomerase from Escherichia coli as biocatalyst for tagatose production. World Journal of Microbiology and Biotechnology, 19, 47–51.
Seo, M. J. (2013). Characterization of an l-arabinose isomerase from Bacillus thermoglucosidasius for d-tagatose production. Bioscience, Biotechnology, and Biochemistry, 77, 385–388.
Hung, X. G., Tseng, W. C., Liu, S. M., & Tzou, W. S. (2014). Characterization of a thermophilic l-arabinose isomerase from Thermoanaerobacterium saccharolyticum NTOU1. Biochemical Engineering Journal, 83, 121–128.
Kim, J. H., Prabhu, P., Jeya, M., Tiwari, M. K., Moon, H. J., Singh, R. K., & Lee, J. K. (2010). Characterization of an l-arabinose isomerase from Bacillus subtilis. Applied Microbiology and Biotechnology, 85, 1839–1847.
Zhan, Y., Xu, Z., Li, S., Liu, X., Xu, L., Feng, X., & Xu, H. (2014). Coexpression of β-d-galactosidase and l-arabinose isomerase in the production of d-tagatose: A functional sweetener. Journal of Agricultural and Food Chemistry, 62, 2412–2417.
Rhimi, M., Bajic, G., Ilhammami, R., Boudebbouze, S., Maguin, E., Haser, R., & Aghajari, N. (2011). The acid-tolerant l-arabinose isomerase from the mesophilic Shewanella sp. ANA-3 is highly active at low temperatures. Microbial Cell Factories, 10, 96–107.
Zhang, H., Jiang, B., & Pan, B. (2007). Purification and characterization of l-arabinose isomerase from Lactobacillus plantarum producing d-tagatose. World Journal of Microbiology and Biotechnology, 23, 641–646.
Acknowledgements
This study was partially sponsored with funds of the Projects CAI + D 2016 50420150100051 LI (Universidad Nacional del Litoral, Santa Fe, Argentina), PIP 2015–2017 No. 11220150100606CO (CONICET, Buenos Aires, Argentina), PICT-201-0249 (Agencia Nacional de Promoción Científica y Tecnológica, Buenos Aires, Argentina). Also, the authors are grateful for the financial support provided by the Brazilian research agencies CAPES, CNPq (407363/2013-0) and FUNCAP (Project Number PRONEX PR2-0101-00012.01.00/15).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Manzo, R.M., Antunes, A.S.L.M., de Sousa Mendes, J. et al. Biochemical Characterization of Heat-Tolerant Recombinant l-Arabinose Isomerase from Enterococcus faecium DBFIQ E36 Strain with Feasible Applications in d-Tagatose Production. Mol Biotechnol 61, 385–399 (2019). https://doi.org/10.1007/s12033-019-00161-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-019-00161-x