Abstract
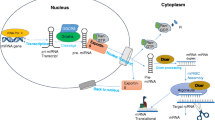
MicroRNA (miRNA) is a small section of ribonucleic acid (RNA) that reduces the protein formation by making the pair of the complementary piece of mRNA. The genes of miRNA are present as transcriptional or polycistronic units in the chromosomes. The cellular multiplication, separation and existence like the multitude of genetic functions are affected by miRNA. Nearly 50% of identified miRNA are located in the residence in the intronic part of the genes. The mature miRNA is yielded in two steps. Drosha and RNA-induced silencing complex are the catalysts that play an important role in miRNA synthesis. The miRNA may function by just hindering the translation or complete vitiation of miRNA that occurs to control the genes. The microRNA antagonists and miRNA mimics are therapeutics approaches for the treatment of abnormalities. The upregulation and downregulation of miRNAs are linked to a number of diseases as miR-122 is associated with viral hepatitis, and some members of let-7 and other miRNAs are concerned with various diseases. Overexpressed miRNAs may function as both oncogenes and regulator of cellular processes. The miRNA functions can be altered by single-point mutations in miRNA target and epigenetic silencing of transcription units. There are numerous molecular targets for miRNA as degradation by nuclease and phosphodiesterase. Thus, miRNA has potential applications in disease diagnosis along with therapy, but the mechanisms involved in miRNA systems and its targeted delivery of miRNA are much more important to achieve its therapeutic applications.
Similar content being viewed by others
References
Gu, W., et al. (2013). Biological basis of miRNA action when their targets are located in human protein-coding region. PLoS ONE, 8(2013), 63403.
MacFarlane, L. A., & Murphy, P. R. (2010). MicroRNA: Biogenesis. Function, and Role in CancerCurr Genomics, 11(2010), 537–561.
Schirle, N. T., et al. (2014). Structural basis for microRNA targeting. Science, 346, 608–613.
Korf, I. (2013). Genomics: The state of the art in RNA-seq analysis. Nature Methods, 10, 1165–1166.
Geeleher, P., et al. (2012). The regulatory effect of miRNAs is a heritable genetic trait in humans. BMC Genomics, 13, 383.
Kuchen, S., et al. (2010). Regulation of MicroRNA expression and abundance during lymphopoiesis. Immunity, 32, 828–839.
Orang, A. V., Safaralizadeh, R., & Kazemzadeh-Bavili, M. (2014). Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. International Journal of Genomics. doi:10.1155/2014/970607.
Winter, J., & Diederichs, S. (2011). MicroRNA biogenesis and cancer. Methods in Molecular Biology, 676, 3–22.
Melamed, Z., et al. (2013). Alternative splicing regulates biogenesis of miRNAs located across exon–intron junctions. Molecular Cell, 50, 869–881.
Melo, C. A., & Melo, S. A. (2014). Non-coding RNAs and Cancer. In Biogenesis and physiology of microRNAs (Vol. 2, pp. 5–24). Springer Science and business media.
Chen, K., & Rajewsky, N. (2007). The evolution of gene regulation by transcription factors and microRNAs. Nature Reviews Genetics, 8, 93–103.
Filipowicz, W., et al. (2008). Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nature Reviews Genetics, 9(2008), 102–114.
Ctalanotto, C., et al. (2016). MicroRNA in control of gene expression: An overview of nuclear functions. International Journal of Molecular Sciences, 17, 1712.
Tamir, L. A., et al. (2014). The interplay between pre-mRNA splicing and microRNA biogenesis within the supra spliceosome. Nucleic Acids Research, 42(7), 4640–4651.
Katahira, J., & Yoneda, Y. (2011). Nucleocytoplasmic transport of MicroRNAs and related small RNAs. Traffic, 12, 1468–1474.
Shomron, N., & Levy, C. (2009). MicroRNA-biogenesis and Pre-mRNA splicing crosstalk (pp. 1–6). doi:10.1155/2009/594678.
Czech, B., & Hannon, G. J. (2011). Small RNA sorting: Matchmaking for Argonautes. Nature Reviews Genetics, 12, 19–31.
Wahid, F., et al. (1803). MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochimica et Biophysica Acta, 2010, 1231–1243.
Rooij, E. V. (2011). The art of MicroRNA research. Circulation Research, 108(2011), 219–234.
Lee, S. J., et al. (2011). Selective nuclear export mechanism of small RNAs. Current Opinion in Structural Biology, 2, 101–108.
Bartel, D. P. (2004). MicroRNAs: Genomics. Biogenesis, Mechanism, and Function, Cell, 166, 281–297.
Lu, Y. C., & Cheng, A. J. (2014). Pathological function and clinical significance of microRNA-10b in cancer. Cancer Science and Research is an Open Access, 1, 1–5.
Jin, H. Y., & Xiao, C. (2015). MicroRNA mechanisms of action: What have we learned from Mice? Frontiers in Genetics, 6, 328.
Carthew, R. W., & Sontheimer, E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell, 136, 642–655.
Wilczynska, A., & Bushell, M. (2015). The complexity of miRNA-mediated repression. Cell Death and Differentiation, 22, 22–33.
Nilsen, T. W. (2007). Mechanisms of microRNA-mediated gene regulation in animal cells. Trends in Genetics, 23(2007), 243–249.
Selbach, M., et al. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature, 455, 58–63.
Dalmay, T. (2013). Mechanism of miRNA-mediated repression of mRNA translation. Biochemical Society Essays in Biochemistry, 54, 29–38.
Pillai, R. S., et al. (2007). Repression of protein synthesis by miRNAs: How many mechanisms? Trends in Cell Biology, 17, 118–126.
Blahna, M. T., & Hata, A. (2012). Smad-mediated regulation of microRNA biosynthesis. FEBS Letters, 586, 1906–1912.
Xie, Z., et al. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Current Biology, 13, 784–789.
Bagga, S., et al. (2005). Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell, 122, 553–563.
Key, M. J., & Gonçalve, I. C. (2014). The role of miRNA in motorneuron disease. Frontiers in Cellular Neuroscience, 8, 1–8.
Ardekani, A. M., & Naeini, M. M. (2010). The role of MicroRNAs in human diseases. Avicenna Journal of Medical Biotechnology, 2(2010), 161–179.
Cristopher, A. F., et al. (2016). MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspectives in Clinical Research, 7, 68–77.
Girard, M., Jacquemin, E., Munnich, A., et al. (2008). miR-122, a paradigm for the role of microRNAs in the liver. Journal of Hepatology, 48, 648–656.
Hou, W., Tian, Q., Zheng, J., et al. (2010). MicroRNA-196 represses Bach1 protein and hepatitis C virus gene expression in human hepatoma cells expressing hepatitis C viral proteins. Hepatology, 51, 1494–1504.
Xu, H., He, J., Dong Xiao, Z., et al. (2010). Liver-enriched transcription factors regulate MicroRNA-122 that targets CUTL1 during liver development. Hepatology, 52, 1431–1442.
Geng Deng, X., Lin Qiu, R., & Hao Wu, Y. (2014). Overexpression of miR-122 promotes the hepatic differentiation and maturation of mouse ESCs through a miR-122/FoxA1/HNF4a-positive feedback loop. Liver International, 34, 281–295.
Coulouarn, C., Factor, V. M., Andersen, J. B., et al. (2009). Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene, 28, 3526–3536.
Zeisel, M. B., Pfeffer, S., Baumert, T. F., et al. (2013). miR-122 acts as a tumor suppressor in hepatocarcinogenesis in vivo. Hepatology, 58, 821–823.
Jangra, R. K., Yi, M., & Lemon, S. M. (2010). Regulation of hepatitis C virus translation and infectious virus production by the MicroRNA miR-122. Journal of Virology, 84, 6615–6625.
Inga Henke, J., Goergen, D., Zheng, J., et al. (2008). microRNA-122 stimulates translation of hepatitis C virus RNA. The EMBO Journal, 27, 3300–3310.
Li, G., & Xiong, X. (2009). MicroRNAs and hepatitis viruses. Frontiers of Medicine in China, 3, 265–270.
Marquart, T. J., Allen, R. M., Ory, D. S., et al. (2010). miR-33 links SREBP-2 induction to repression of sterol transporters. Proceedings of the National Academy of Sciences. doi:10.1073/pnas.1005191107.
Rayner, K. J., Suarez, Y., Davalos, A., et al. (2010). MiR-33 contributes to the regulation of cholesterol homeostasis. Science, 328, 1570–1573.
Krutzfeldt, J., & Stoffe, M. (2006). MicroRNAs: A new class of regulatory genes affecting metabolism. Cell Press, 4(1), 9–12.
Connel, R. M. O., Taganov, K. D., Boldin, M. P., et al. (2007). MicroRNA-155 is induced during the macrophage inflammatory response. Immunology, 104, 1604–1609.
Dorsett, Y., Mcbride, K. M., Jankovic, M., et al. (2008). MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity, 28, 630–638.
Xiao, C., & Rajewsky, K. (2009). MicroRNA control in the immune system: Basic principles. Cell, 136(2009), 26–36.
Li, Y., Guessos, F., Zhang, Y., et al. (2010). MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Research, 69, 7569–7576.
Zhang, Y., et al. (2012). The role of microRNAs in glioma initiation and progression. Frontiers in Bioscience, 17, 700–712.
Lou, J. W., et al. (2015). Role of micro-RNA (miRNA) in the pathogenesis of glioblastoma. European Review for Medical and Pharmacological Sciences, 19, 1630–1639.
Feinberg, M. W., & Moore, K. J. (2016). MicroRNA regulation of atherosclerosis. Circulation Research, 118, 703–720.
Boon, R., & Vickers, K. C. (2013). Intercellular transport of MicroRNA. Arteriosclerosis, Thrombosis, and Vascular Biology, 33, 186–192.
Libanio, D., et al. (2015). Helicobacter pylori and microRNAs: Relation with innate immunity and progression of preneoplastic conditions. World Journal of Clinical Oncology, 10(6), 111–132.
Eulalio, A., et al. (2012). The mammalian microRNA response to bacterial infections. RNA Biology, 9, 742–750.
Schulte, L. N., et al. (2012). Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. International Journal of Molecular Sciences, 13, 1173–1185.
Izar, B., et al. (2017). microRNA response to Listeria monocytogenes infection in epithelial. Cells, Frontiers in Immunology, 8, 107.
Salama, A., et al. (2014). MicroRNA-29b modulates innate and antigen-specific immune responses in mouse models of autoimmunity. PLoS ONE, 9, e106153.
Ahluwalia, P. K., et al. (2013). Perturbed microRNA expression by mycobacterium tuberculosis promotes macrophage polarization leading to pro-survival foam cell.
Bandyopadhyay, S., Long, M. E., & Allen, L. A. H. (2014). Differential expression of microRNAs in Francisella tularensis-infected human macrophages: miR-155-dependent downregulation of MyD88 inhibits the inflammatory response. PLoS ONE, 9(10), e109525. doi:10.1371/journal.pone.0109525.
Skalsky, R. L., & Cullen, B. R. (2010). Viruses, microRNAs, and host interactions. Annual Review of Microbiology, 64(2010), 123–141.
Broderick, J. A., & Zamore, P. D. (2011). MicroRNA therapeutics. Gene Therapy, 18, 1104–1110.
Jackson, A. I., & Linsley, P. S. (2010). The therapeutic potential of microRNA modulation. Discovery Medicine, 9(2010), 311–318.
Li, Z., & Rana, T. M. (2014). Therapeutic targeting of microRNAs: Current status and future challenges. Nature Reviews Drug Discovery, 13(2014), 622–638.
Leung, A. K. L., & Sharp, P. A. (2010). MicroRNA functions in stress responses. Molecular Cell, 22(40), 205–215.
Stenvang, J., Petri, A., Lindow, M., Obad, S., & Kauppinen, S. (2012). Inhibition of microRNA function by antimiR oligonucleotides. Silence, 3, 1. doi:10.1186/1758-907X-3-1.
Rooji, E. V., & Kauppinen, S. (2014). Development of microRNA therapeutics is coming of age. EMBO Molecular Medicine, 6, 851–864.
Jackson, A., & Linsley, P. S. (2010). The Therapeutic Potential of microRNA Modulation. Molecular Cell, 40, 205–215.
Liu, J., et al. (2005). MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nature Cell Biology, 7(2005), 719–723.
Nedaeinia, R., et al. (2017). Current status and perspectives regarding LNA-anti-miR oligonucleotides and microRNA miR-21 inhibitors as a potential therapeutic option in treatment of colorectal cancer. Journal of Cellular Biochemistry. doi:10.1002/jcb.26047.
Obad, S., et al. (2011). Silencing of microRNA families by seed-targeting tiny LNAs. Nature Genetics, 43, 371–378.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Naveed, A., ur-Rahman, S., Abdullah, S. et al. A Concise Review of MicroRNA Exploring the Insights of MicroRNA Regulations in Bacterial, Viral and Metabolic Diseases. Mol Biotechnol 59, 518–529 (2017). https://doi.org/10.1007/s12033-017-0034-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-017-0034-7