Abstract
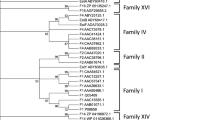
A gene encoding a carboxylesterase produced by Geobacillus thermoleovoras CCR11 was cloned in the pET-3b cloning vector, sequenced and expressed in Escherichia coli BL21(DE3). Gene sequence analysis revealed an open reading frame of 750 bp that encodes a polypeptide of 250 amino acid residues (27.3 kDa) named CaesCCR11. The enzyme showed its maximum activity at 50 °C and pH 5–8, with preference for C4 substrates, confirming its esterase nature. It displayed good resistance to temperature, pH, and the presence of organic solvents and detergents, that makes this enzyme biotechnologically applicable in the industries such as fine and oleo-chemicals, cosmetics, pharmaceuticals, organic synthesis, biodiesel production, detergents, and food industries. A 3D model of CaesCCR11 was predicted using the Bacillus sp. monoacyl glycerol lipase bMGL H-257 structure as template (PBD code 3RM3, 99 % residue identity with CaesCCR11). Based on its canonical α/β hydrolase fold composed of 7 β-strands and 6 α-helices, the α/β architecture of the cap domain, the GLSTG pentapeptide, and the formation of distinctive salt bridges, we are proposing CaesCCR11 as a new member of family XV of lipolytic enzymes.





Similar content being viewed by others
References
Sharma, R., Chisti, Y., & Banerjee, U. C. (2001). Production, purification, characterization, and applications of lipases. Biotechnology Advances, 19, 627–662.
Gupta, R., Kumari, A., Syal, P., & Singh, Y. (2015). Molecular and functional diversity of yeast and fungal lipases: their role in biotechnology and cellular physiology. Progress in Lipid Research, 57, 40–54.
Bornscheuer, U. T. (2002). Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiology Reviews, 26, 73–81.
Soliman, N. A., Knoll, M., Abdel-Fattah, Y. R., Schmid, R. D., & Lange, S. (2007). Molecular cloning and characterization of thermostable esterase and lipase from Geobacillus thermoleovorans YN isolated from desert soil in Egypt. Process Biochemistry, 42, 1090–1100.
Arpigny, J. L., & Jaeger, K. E. (1999). Bacterial lipolytic enzymes: classification and properties. Biochemical Journal, 343(Pt 1), 177–183.
Montoro-García, S., Martínez-Martínez, I., Navarro-Fernández, J., Takami, H., García-Carmona, F., & Sánchez-Ferrer, A. (2009). Characterization of a novel thermostable carboxylesterase from Geobacillus kaustophilus HTA426 shows the existence of a new carboxylesterase family. Journal of Bacteriology, 191, 3076–3085.
Charbonneau, D., Meddeb-Mouelhi, F., & Beauregard, M. (2010). A novel thermostable carboxylestarese from Geobacillus thermodenitrificans: evidence for a new carboxylesterase family. Journal of Biochemistry, 148, 299–308.
Rengachari, S., Bezerra, G. A., Riegler-Berket, L., Gruber, C. C., Sturm, C., Taschler, U., et al. (2012). The structure of monoacylglycerol lipase from Bacillus sp. H-257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes. Biochimica et Biophysica Acta, 1821, 1012–1021.
Charbonneau, D. M., & Beauregard, M. (2013). Role of key salt bridges in thermostability of G. thermodenitrificans EstGtA2: distinctive patterns within the new bacterial lipolytic enzyme family XV. PLoS One, 10(8), e0136940. doi:10.1371/journal.pone.0136940.
Rao, L., Xue, Y., Zheng, Y., Lu, J. R., & Ma, Y. (2013). A novel alkaliphilic Bacillus 324 esterase belongs to the 13th bacterial lipolytic enzyme family. PLoS One, 8, e60645. doi:10.1371/journal.pone.0060645.
Castro-Ochoa, L. D., Rodríguez-Gómez, C., Valerio-Alfaro, G., & Oliart-Ros, R. (2005). Screening, purification and characterization of the thermoalkalophilic lipase produced by Bacillus thermoleovorans CCR11. Enz. Microb. Technol., 37, 648–654.
Quintana-Castro, R., Díaz, P., Valerio-Alfaro, G., García, H. S., & Oliart-Ros, R. (2009). Gene cloning, expression and characterization of the Geobacillus thermoleovorans CCR11 thermoalkaliphilic lipase. Molecular Biotechnology, 42, 75–83.
Sánchez-Otero, M. G., Ruiz-López, I. I., Avila-Nieto, D. E., & Oliart-Ros, R. M. (2011). Significant improvement of Geobacillus thermoleovorans CCR11 thermoalkalophilic lipase production using response surface methodology. New Biotechnology, 28, 761–766.
Schmidt-Dannert, C., Rúa, L., Atomi, H., & Schmid, R. (1996). Thermoalkalophilic lipase of Bacillus thermocatenulatus. I. Molecular cloning, nucleotide sequence, purification and some properties. Biochimica et Biophysica Acta, 1301, 105–114.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Prim, N., Sánchez, M. M., Ruiz, C., Pastor, F. I. J., & Díaz, P. (2003). Use of methylumbeliferyl-derivative substrates for lipase activity characterization. Journal of Molecular Catalysis B: Enzymatic, 22, 339–346.
Lowry, O. H., Rosebrough, N. J., Farr, A. L., & Randall, R. J. (1951). Protein measurement with the Folin phenol reagent. Journal of Biological Chemistry, 193, 265–275.
Nawani, N., Dosanjh, S., & Kaur, J. (1998). A novel thermostable lipase from a thermophilic Bacillus sp.: characterization and esterification studies. Biotechnology Letters, 20, 997–1000.
Ausubel, F., Brent, R. R., Kingstone, M. D., Seidman, S. J., & Struhl, K. (Eds.) (1997). Short protocols in molecular biology. A compendium of methods from current protocols in molecular biology. Vol. I Preparation and analysis of DNA and enzymatic manipulation of DNA (pp. 2-1–3-50) New York: Wiley.
Maniatis, T., Fritsch, E., & Sambrook, J. (1982). Molecular Cloning: A laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
Grossman, T. H., Kawasaki, E. S., Punreddy, S. R., & Osburne, M. S. (1998). Spontaneous cAMP-dependent derepression of gene expression in stationary phase plays a role in recombinant expression instability. Gene, 209, 95–103.
Kalendar, R., Lee, D., & Schulman, A. H. (2009). FastPCR Software for PCR primer and probe design and repeat search genes: Focus on bioinformatics. Genes, Genomes and Genomics, 3(1), 1–14.
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., & Bairoch, A. (2005). Protein identification and analysis tools on the ExPASy Server. In J. M. Walker (Ed.), The proteomics protocols handbook (pp. 571–607). Totowa: Humana Press.
Rost, R., Yachdav, G., & Liu, J. (2004). The predict protein server. Nucleic Acids Research, 32(Web Server issue), W321–W326.
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position, specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729.
Robert, X., & Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research, 42(Web Server issue), W320–W324.
Källberg, M., Wang, H., Wang, S., Peng, J., Wang, Z., Lu, H., & Xu, J. (2012). Template-based protein structure modeling using the RaptorX web server. Nature Protocols, 7, 1511–1522.
Constantini, S., Colonna, G., & Facchiano, A. M. (2008). ESBRI: A web server for evaluating salt bridges in proteins. Bioinformation, 3, 137–138.
Takami, H., Takaki, Y., Chee, G. J., Nishi, S., Shimamura, S., Suzuki, H., et al. (2004). Thermoadaptation trait revealed by the genome sequence of thermophilic Geobacillus kaustophilus. Nucleic Acids Research, 32, 6292–6303.
Rúa, M. L., Schmidt-Dannert, C., Wahl, S., Sprauer, A., & Schmid, R. D. (1997). Thermoalkalophilic lipase of Bacillus thermocatenulatus large-scale production, purification and properties: aggregation behavior and its effect on activity. Journal of Biotechnology, 56, 89–102.
Salameh, M. A., & Wiegel, J. (2010). Effects of detergents on activity, thermostability and aggregation of two alkalithermophilic lipases from Thermosyntropha lipolytica. Open Biochemistry Journal, 4, 22–28.
Schlieben, N. H., Niefind, K., & Schomburg, D. (2004). Expression, purification and aggregation studies of His-tagged thermoalkalophilic lipase from Bacillus thermocatenolatus. Protein Expression and Purification, 34, 103–110.
Imamura, S., & Kitaura, S. (2000). Purification and characterization of a monoacylglycerol lipase from the moderately thermophilic Bacillus sp. H-257. Journal of Biochemistry, 127, 419–425.
Kitaura, S., Suzuki, K., & Imamura, S. (2001). Monoacylglycerol lipase from moderately thermophilic Bacillus sp. strain H-257: molecular cloning, sequencing, and expression in Escherichia coli of the gene. Journal of Biochemistry, 129, 397–402.
Zhu, Y., Lid, J., Caib, H., Nib, H., Xiaob, A., & Hou, L. (2013). Characterization of a new and thermostable esterase from a metagenomic library. Microb. Res., 168, 589–597.
Ceroni, A., Passerini, A., Vullo, A., & Frasconi, P. (2006). DISULFIND: A disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Research, 34(1), W177–W181.
Boonmak, C., Takahasi, Y., & Morikawa, M. (2013). Draft genome sequence of Geobacillus thermoleovorans strain B23. Genome Announcements, 1(6), e00944-13. doi:10.1128/genomeA.00944-13.
Rengachari, S., Aschauer, P., Schittmayer, M., Mayer, N., Gruber, K., Breinbauer, R., et al. (2013). Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase. Journal of Biological Chemistry, 288, 31093–31104.
Lee, D. W., Kim, H. W., Lee, K. W., Kim, B. C., Choe, E. A., Lee, H. S., et al. (2001). Purification and characterization of two distinct thermostable lipases from the gram-positive thermophilic bacterium Bacillus thermoleovorans ID-1. Enzyme and Microbial Technology, 29, 363–371.
Acknowledgments
M.Sc. Graciela Espinosa Luna and M.Sc. Rodrigo E. Matus Toledo acknowledge their scholarships from the National Council for Science and Technology (Conacyt). This work was supported by Grants No. 105636 from Conacyt, 362.06-P from the General Direction of Superior Technological Education (DGEST-SEP) and PROMEP.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Espinosa-Luna, G., Sánchez-Otero, M.G., Quintana-Castro, R. et al. Gene Cloning and Characterization of the Geobacillus thermoleovorans CCR11 Carboxylesterase CaesCCR11, a New Member of Family XV. Mol Biotechnol 58, 37–46 (2016). https://doi.org/10.1007/s12033-015-9901-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-015-9901-2