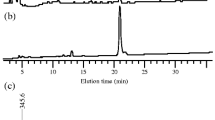
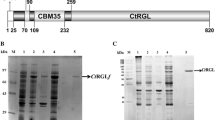
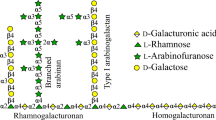
Abstract
Rhamnogalacturonan lyase (PcRGL4A) was purified from the culture supernatant of Penicillium chrysogenum 31B. PcRGL4A optimal activity occurred between pH 7–8 and at 40 °C. Conserved Domain Search analysis identified PcRGL4A as a member of Polysaccharide Lyase family 4. PcRGL4A contains two conserved catalytic and four conserved substrate-binding residues as determined by X-ray crystallography of the Aspergillus aculeatus RG lyase. Recombinant PcRGL4A (rPcRGL4A) expressed in Escherichia coli demonstrated specific activity against rhamnogalacturonan (RG) but not homogalacturonan. Analysis of the RG reaction products by high-performance anion-exchange chromatography revealed that rPcRGL4A cleaved the substrate in an endo-manner and that the major final product was an RG tetrasaccharide with 4-deoxy-4,5-unsaturated galacturonic acid at the nonreducing end. Based on these results, PcRGL4A was classified as an endo-acting RG lyase (EC 4.2.2.23). Divalent cations were not essential for the enzymatic activity of rPcRGL4A, but addition of calcium ions to the reaction mixture increased enzymatic activity. rPcRGL4A demonstrated a preference for RG lacking galactose decoration.






Similar content being viewed by others
References
McNeil, M., Darvill, A. G., Fry, S. C., & Albersheim, P. (1984). Structure and function of the primary cell walls of plants. Annual Review of Biochemistry, 53, 625–663.
Thakur, B. R., Singh, R. K., & Handa, A. K. (1997). Chemistry and uses of pectin—A review. Critical Reviews in Food Science and Nutrition, 37, 47–73.
Willats, W. G. T., McCartney, L., Mackie, W., & Knox, J. P. (2001). Pectin: Cell biology and prospects for functional analysis. Plant Molecular Biology, 47, 9–27.
Voragen, A. G. J., Coenen, G. J., Verhoef, R. P., & Schols, H. (2009). Pectin, a versatile polysaccharide present in plant cell walls. Structural Chemistry, 20, 263–275.
O’Neil, M. A., Warrenfeltz, D., Kates, K., Pellerin, P., Doco, T., Darvill, A. G., et al. (1996). Rhamnogalacturonan-II, a pectic polysaccharide in the walls of growing plant cell, forms a dimer that is covalently cross-linked by a borate ester. In vitro conditions for the formation and hydrolysis of the dimer. Journal of Biological Chemistry, 271, 22923–22930.
Willats, W. G. T., Knox, P., & Mikkelsen, J. D. (2006). Pectin: New insights into an old polymer are starting to gel. Trends in Food Science & Technology, 17, 97–104.
Thomassen, L. V., Vigsnæs, L. K., Licht, T. R., Mikkelsen, J. D., & Meyer, A. S. (2011). Maximal release of highly bifidogenic soluble dietary fibers from industrial potato pulp by minimal enzymatic treatment. Applied Microbiology and Biotechnology, 90, 873–884.
Onumpai, C., Kolida, S., Bonnin, E., & Rastall, R. A. (2011). Microbial utilization and selectivity of pectin fractions with various structures. Applied and Environment Microbiology, 77, 5747–5754.
Lærke, H. N., Meyer, A. S., Kaack, K. V., & Larsen, T. (2007). Soluble fiber extracted from potato pulp is highly fermentable but has no effect on risk markers of diabetes and cardiovascular disease in Goto-Kakizaki rats. Nutrition Research, 27, 152–160.
Schols, H. A., Geraeds, C. C. J. M., Searle-van Leeuwen, M. F., Kormelink, F. J. M., & Voragen, A. G. J. (1990). Rhamnogalacturonase: a novel enzyme that degrades the hairy regions of pectins. Carbohydrate Research, 206, 105–115.
Mutter, M., Colquhoun, I. J., Schols, H. A., Beldman, G., & Voragen, A. G. J. (1996). Rhamnogalacturonase B from Aspergillus aculeatus is a rhamnogalacturonan α-L-rhamnopyranosyl-(1->4)-α-d-galactopyranosyluronide lyase. Plant Physiology, 110, 73–77.
Mutter, M., Colquhoun, I. J., Beldman, G., Schols, H. A., Bakx, E. J., & Voragen, A. G. J. (1998). Characterization of recombinant rhamnogalacturonan α-L-rhamnopyranosyl-(1,4)-α-D-galactopyranosyluronide lyase from Aspergillus aculeatus. An enzyme that fragments rhamnogalacturonan I regions of pectin. Plant Physiology, 117, 141–152.
Mutter, M., Renard, C. M. G. C., Beldman, G., Schols, H. A., & Voragen, A. G. J. (1998). Mode of action of RG-hydrolase and RG-lyase toward rhamnogalacturonan oligomers. Characterization of degradation products using RG-rhamnohydrolase and RG-galacturonohydrolase. Carbohydrate Research, 311, 155–164.
Kofod, L. V., Kauppinen, S., Christgau, S., Andersen, L. N., Heldt-Hansen, H. P., Dörreich, K., et al. (1994). Cloning and characterization of two structurally and functionally divergent rhamnogalacturonases from Aspergillus aculeatus. Journal of Biological Chemistry, 269, 29182–29189.
Laatu, M., & Condemine, G. (2003). Rhamnogalacturonate lyase RhiE is secreted by the out system in Erwinia chrysanthemi. Journal of Bacteriology, 185, 1642–1649.
Yoshino-Yasuda, S., Karita, S., Kato, M., & Kitamoto, N. (2012). Sequence analysis and heterologous expression of rhamnogalacturonan lyase A gene (AsrglA) from Shoyu Koji Mold, Aspergillus sojae KBN1340. Food Science and Technology Research, 18, 901–909.
McKie, V. A., Vincken, J. P., van den Voragen, A. G. J., Broek, L. A. M., Stimson, E., & Gilbert, H. J. (2001). A new family of rhamnogalacturonan lyases contains an enzyme that binds to cellulose. Biochemical Journal, 355, 167–177.
Pagès, S., Valette, O., Abdou, L., Bélaïch, A., & Bélaïch, J. P. (2003). A rhamnogalacturonan lyase in the Clostridium cellulolyticum cellulosome. Journal of Bacteriology, 185, 4727–4733.
Ochiai, A., Yamasaki, M., Itoh, T., Mikami, B., Hashimoto, W., & Murata, K. (2006). Crystallization and preliminary X-ray analysis of the rhamnogalacturonan lyase YesW from Bacillus subtilis strain 168, a member of polysaccharide lyase family 11. Acta Crystallographica Section F, 62, 438–440.
Ochiai, A., Itoh, T., Kawamata, A., Hashimoto, W., & Murata, K. (2007). Plant cell wall degradation by saprophytic Bacillus subtilis strains: gene clusters responsible for rhamnogalacturonan depolymerization. Applied and Environment Microbiology, 73, 3803–3813.
Ochiai, A., Itoh, T., Maruyama, Y., Kawamata, A., Mikami, B., Hashimoto, W., et al. (2007). A novel structural fold in polysaccharide lyases: Bacillus subtilis family 11 rhamnogalacturonan lyase YesW with an eight-bladed β-propeller. Journal of Biological Chemistry, 282, 37134–37145.
Silva, I. R., Larsen, D. M., Meyer, A. S., & Mikkelsen, J. D. (2011). Identification, expression, and characterization of a novel bacterial RGI lyase enzyme for the production of bio-functional fibers. Enyzme and Microbial Technology, 49, 160–166.
Sakamoto, T., & Thibault, J. F. (2001). An exo-arabinanase of Penicillium chrysogenum able to release arabinobiose from α-1,5-L-arabinan. Applied and Environment Microbiology, 67, 3319–3321.
Sakamoto, T., Ihara, H., Kozaki, S., & Kawasaki, H. (2003). A cold-adapted endo-arabinanase from Penicillium chrysogenum. Biochimica et Biophysica Acta, 1624, 70–75.
Sakamoto, T., Ihara, H., Shibano, A., Kasai, N., Inui, H., & Kawasaki, H. (2004). Molecular characterization of a Penicillium chrysogenum exo-1,5-α-L-arabinanase that is structurally distinct from other arabinan-degrading enzymes. FEBS Letters, 560, 199–204.
Sakamoto, T., Ogura, A., Inui, M., Tokuda, S., Hosokawa, S., Ihara, H., et al. (2011). Identification of a GH62 α-L-arabinofuranosidase specific for arabinoxylan produced by Penicillium chrysogenum. Applied Microbiology and Biotechnology, 90, 137–146.
Sakamoto, T., Inui, M., Yasui, K., Tokuda, S., Akiyoshi, M., Kobori, Y., et al. (2012). Biochemical characterization and gene expression of two endo-arabinanases from Penicillium chrysogenum 31B. Applied Microbiology and Biotechnology, 93, 1087–1096.
Sakamoto, T., Inui, M., Yasui, K., Hosokawa, S., & Ihara, H. (2013). Substrate specificity and gene expression of two Penicillium chrysogenum α-L-arabinofuranosidases (AFQ1 and AFS1) belonging to glycoside hydrolase families 51 and 54. Applied Microbiology and Biotechnology, 97, 1121–1130.
Shinozaki, A., Kawakami, T., Hosokawa, S., & Sakamoto, T. (2014). A novel GH43 α-L-arabinofuranosidase of Penicillium chrysogenum that preferentially degrades single-substituted arabinosyl side chains in arabinan. Enyzme and Microbial Technology, 58–59, 80–86.
Thibault, J. F., Renard, C. M. G. C., Axelos, M. A. V., Roger, P., & Crépeau, M. J. (1993). Studies of the length of homogalacturonic regions in pectins by acid hydrolysis. Carbohydrate Research, 238, 271–286.
Tanabe, H., Kobayashi, Y., Matuo, Y., Nishi, N., & Wada, F. (1984). Isolation and fundamental properties of endo-pectate lyase pI-isozymes from Erwinia carotovora. Agricultural and Biological Chemistry, 48, 2113–2120.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Shevchenko, A., Wilm, M., Vorm, O., & Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical Chemistry, 68, 850–858.
Blumenkrantz, N., & Asboe-Hansen, G. (1973). New method for quantitative determination of uronic acids. Analytical Biochemistry, 54, 484–489.
Van den Berg, M. A., Albang, R., Albermann, K., Badger, J. H., Daran, J. M., Driessen, A. J., et al. (2008). Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nature Biotechnology, 26, 1161–1168.
Marcet-Houben, M., de la Ballester, A. R., Fuente, B., Harries, E., Marcos, J. F., González-Candelas, L., et al. (2012). Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genomics, 13, 646.
Galagan, J. E., Calvo, S. E., Cuomo, C., Ma, L.-J., Wortman, J. R., Batzoglou, S., et al. (2005). Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature, 438, 1105–1115.
McDonough, M. A., Kadirvelraj, R., Harris, P., Poulsen, J. C., & Larsen, S. (2004). Rhamnogalacturonan lyase reveals a unique three-domain modular structure for polysaccharide lyase family 4. FEBS Letters, 565, 188–194.
Jensen, M. H., Otten, H., Christensen, U., Borchert, T. V., Christensen, L. L. H., Larsen, S., et al. (2010). Structural and biochemical studies elucidate the mechanism of rhamnogalacturonan lyase from Aspergillus aculeatus. Journal of Molecular Biology, 404, 100–111.
Wong, D. (2008). Enzymatic deconstruction of backbone structures of the ramified regions in pectins. Protein Journal, 27, 30–42.
Bonnin, E., Garnier, C., & Ralet, M. C. (2014). Pectin-modifying enzymes and pectin-derived materials: applications and impacts. Applied Microbiology and Biotechnology, 98, 519–532.
Acknowledgments
This work was supported by JSPS KAKENHI Grant Number 25450135.
Author information
Authors and Affiliations
Corresponding author
Additional information
M. Iwai and H. Yamada are equal first authors.
Rights and permissions
About this article
Cite this article
Iwai, M., Yamada, H., Ikemoto, T. et al. Biochemical Characterization and Overexpression of an Endo-rhamnogalacturonan Lyase from Penicillium chrysogenum . Mol Biotechnol 57, 539–548 (2015). https://doi.org/10.1007/s12033-015-9847-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-015-9847-4