Abstract
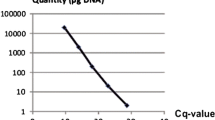
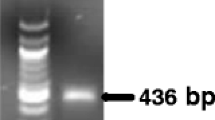
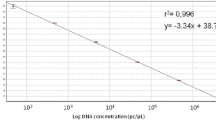
Rice blast caused by Magnaporthe oryzae is a major disease in the paddy field and also a representative model system in the investigation of plant–microbe interactions. This study was undertaken to provide the quantitative evaluation method that specifically determines the amount of M. oryzae proliferation in planta. Real-time PCR was used as the detection strategy in combination with the primer pair and Taqman probe specific to MHP1, a unigene encoding HYDROPHOBIN that is indispensable for normal virulence expression. Based on the crossing point values from the PCR reactions containing a series of increasing concentration of cloned amplicon or fungal genomic DNA, correlation among the template’s copy number or its amount and amplification pattern was calculated. Reliability of this equation was further confirmed using the DNA samples from the rice leaves infected with compatible or incompatible strains of M. oryzae. The primer pair used in the Taqman real-time PCR reaction can recognize the existence of fungal DNA as low as 1 pg. In sum, our quantitative evaluation system is applicable and reliable in the blast diagnosis and also in the estimation of objective blast disease progression.



Similar content being viewed by others
References
Valent, B., & Chumley, F. G. (1991). Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea. Annual Review of Phytopathology, 29, 443–467.
Widawsky, D. A., & O’Toole, J. C. (1990). Prioritizing the rice biotechnology research agenda for eastern India. New York: The Rockefeller Foundation.
Lee, F. N. (1994). Rice breeding programs, blast epidemics and blast management in the United States. In R. S. Zeigler, et al. (Eds.), Rice blast disease (pp. 489–500). Wallingford: CABI and International Rice Research Institute.
Cho, Y. C., Suh, J. P., Jeung, J. U., Roh, J. H., Yang, C. I., Oh, M. K., et al. (2009). Resistance genes and their effect on blast in Korean rice varieties (Oryza sativa L.). In G. L. Wang & B. Valent (Eds.), Advances in genetics, genomics and control of rice blast disease (pp. 291–304). Berlin: Springer.
Endo, T., Nakamura, T., Yonemaru, J., Ishikawa, G., Yamaguchi, M., Kataoka, T., et al. (2009). Genetic analysis of resistance against bacterial leaf blight and leaf blast disease in the Japanese rice cultivar Asominori. In G. L. Wang & B. Valent (Eds.), Advances in genetics, genomics and control of rice blast disease (pp. 291–304). Berlin: Springer.
Sobrizal, S., & Anggiani, S. (2007). Rice blast disease in Indonesia. In JIRCAS Working Report, 53, pp. 71–79.
Howard, R. J., Ferrari, M. A., Roach, D. H., & Money, N. P. (1991). Penetration of hard substrates by a fungus employing enormous turgor pressures. Proceedings of the National Academy of Science of United States of America, 88, 11281–11284.
Heath, M. C., Valent, B., Howard, R. J., & Chumley, F. G. (1990). Correlations between cytologically detected plant-fungal interactions and pathogenicity of Magnaporthe grisea toward weeping lovegrass. Phytopathology, 80, 1382–1386.
Heath, M. C., Howard, R. J., Valent, B., & Chumley, F. G. (1992). Ultrastructural interactions of one strain of Magnaporthe grisea with goosegrass and weeping lovegrass. Canadian Journal of Botany, 70, 779–787.
Tucker, S. L., & Talbot, N. J. (2001). Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annual Review of Phytopathology, 39, 385–401.
Ebbole, D. J. (2007). Magnaporthe as a model for understanding host–pathogen interactions. Annual Review of Phytopathology, 45, 437–456.
Su’udi, M., Kim, M. G., Park, S. R., Hwang, D. J., Bae, S. C., & Ahn, I. P. (2011). Arabidopsis cell death in compatible and incompatible interactions with Alternaria brassicicola. Molecules and Cells, 31, 593–601.
Cho, Y. C., Kwon, S. W., Suh, J. P., Kim, J. J., Lee, J. H., Roh, J. H., et al. (2008). QTLs identification and confirmation of field resistance to leaf blast in temperate japonica rice (Oryza sativa L.). Journal of Crop Science and Biotechnology, 11, 269–276.
Kim, B. R., Roh, J. H., Choi, S. H., Ahn, S. W., & Han, S. S. (2004). Durability of rice cultivars to blast in Korea by sequential planting method. Korean Journal of Breeding, 36, 350–356.
International Rice Research Institute. (1988). Standard Evaluation System for Rice, International Rice Testing Program. Los Banos: International Rice Testing Program.
Narayanasamy, P. (2001). Plant pathogen detection and disease diagnosis. New York: Marcel Dekker Inc.
Skottrup, P., Hearty, S., Frøkiær, H., Leonard, P., Hejgaard, J., O’Kennedy, R., et al. (2007). Detection of fungal spores using a generic surface plasmon resonance immunoassay. Biosensors & Bioelectronics, 22, 2724–2729.
Frederick, R. D., Snyder, K. E., Tooley, P. E., Berthier-Schaad, Y., Peterson, G. B., Bonde, M. R., et al. (2000). Identification and differentiation of Tilletia indica and T. walker using the polymerase chain reaction. Phytopathology, 90, 951–960.
Chadha, S., & Gopalakrishna, T. (2006). Detection of Magnaporthe grisea in infested rice seeds using polymerase chain reaction. Journal of Applied Microbiology, 100, 1147–1153.
Torres-Calzada, C., Tapia-Tussell, R., Quijano-Ramayo, A., Martin-Mex, R., Rojas-Herrera, R., Higuera-Ciapara, I., et al. (2011). A species-specific polymerase chain reaction assay for rapid and sensitive detection of Colletotrichum capsici. Molecular Biotechnology, 49, 48–55.
Lievens, B., Brouwer, M., Vanachter, A. C., Cammue, B. P., & Thomma, B. P. (2006). Real-time PCR for detection and quantification of fungal and oomycete tomato pathogens in plant and soil samples. Plant Science, 171, 155–165.
Selma, M. V., Martínez-Culebras, P. V., & Aznar, R. (2008). Real-time PCR based procedures for detection and quantification of Aspergillus carbonarius in wine grapes. International Journal of Food Microbiology, 122, 126–134.
Borneman, J., & Hartin, R. J. (2000). PCR primers that amplify fungal rRNA genes from environmental samples. Applied and Environmental Microbiology, 66, 4356–4360.
Martin, K. J., & Rygiewicz, P. T. (2005). Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiology, 5, 28.
Sone, T., Fukiya, S., Kodama, M., & Tomita, F. (2000). Molecular structure of rDNA repeat unit in Magnaporthe grisea. Bioscience, Biotechnology, and Biochemistry, 64, 1733–1736.
Kim, S., Ahn, I. P., Rho, H. S., & Lee, Y. H. (2005). MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Molecular Microbiology, 57, 1224–1237.
Dhingra, O. D., & Sinclair, J. B. (1985). Basic plant pathology methods. Boca Raton, FL: CRC Press.
Ahn, I. P., Kim, S., Kang, S., Suh, S. C., & Lee, Y. H. (2005). Rice defense mechanisms against Cochliobolus miyabeanus and Magnaporthe grisea are distinct. Phytopathology, 95, 1248–1255.
Kim, S., Ahn, I. P., Park, C. H., Park, S. G., Park, S. Y., Jwa, N. S., et al. (2001). Molecular characterization of the cDNA encoding an acidic isoform of PR-1 protein in rice. Molecules and Cells, 11, 115–121.
Stewart, C. N., & Via, L. E. (1993). A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. BioTechniques, 14, 748–751.
Feinberg, A. P., & Vogelstein, B. (1983). A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Analytical Biochemistry, 132, 6–13.
Simon, U. K., & Weiß, M. (2008). Intragenomic variation of fungal ribosomal genes is higher than previously thought. Molecular Biology and Evolution, 25, 2251–2254.
Qi, M., & Yang, Y. (2002). Quantification of Magnaporthe grisea during infection of rice plants using real-time PCR and northern blot/phosphoimaging analysis. Phytopathology, 92, 870–876.
Harmon, P. F., Dunkle, L. D., & Latin, R. (2003). A rapid PCR-based method for the detection of Magnaporthe oryzae from infected perennial ryegrass. Plant Disease, 87, 1072–1076.
Jones, J. D. G., & Dangl, J. L. (2006). The plant immune system. Nature, 444, 323–329.
Acknowledgments
This research was supported by grants from the Research Program for Agricultural Science and Technology Development (Project No. PJ008840) and the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center No. PJ008021072011), Rural Development Administration, Korea.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Su’udi, M., Kim, J., Park, JM. et al. Quantification of Rice Blast Disease Progressions Through Taqman Real-Time PCR. Mol Biotechnol 55, 43–48 (2013). https://doi.org/10.1007/s12033-012-9632-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-012-9632-6