Abstract
G-quartets are square planar arrangements of four guanine bases, which can form extraordinarily stable stacks when present in nucleic acid sequences. Such G-quadruplex structures were long regarded as an in vitro phenomenon, but the widespread presence of suitable sequences in genomes and the identification of proteins that stabilize, modify or resolve these nucleic acid structures have provided circumstantial evidence for their physiological relevance. The therapeutic potential of small molecules that can stabilize or disrupt G-quadruplex structures has invigorated the field in recent years. Here we review some of the key observations that support biological functions for G-quadruplex DNA as well as the techniques and tools that have enabled researchers to probe these structures and their interactions with proteins and small molecules.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
More than four decades before Watson and Crick proposed their structure for DNA, the German chemist Ivar Bang noted that guanylic acid forms gels at high millimolar concentrations [1]. This unusual physical property puzzled researchers for the next 50 years until Gellert and colleagues collected fiber X-ray diffraction data on guanylic acid [2], revealing the assembly of tetrameric units into large helical structures that account for the gel-like properties of the aqueous solution. Four molecules of guanylic acid form a square planar arrangement in which each of the four bases is the donor and acceptor of two hydrogen bonds, now referred to as a G-quartet (Fig. 1). As interest in nucleic acids intensified over the following decades, it became clear that guanosine homo-oligomers can adopt the same structure, both in the ribose and deoxyribose forms [3, 4]. For years, little consideration was given to possible roles for G-quartets in biological systems until Henderson and colleagues made the observation that oligonucleotides corresponding to the G-rich strand of telomeric DNA display unexpectedly high electrophoretic mobility on nondenaturing polyacrylamide gels [5]. Structural probing later showed that G-rich sequences found at telomeres and in the immunoglobulin switch region can indeed adopt stable four-stranded structures now known as G-quadruplexes [5–8].
Among the five nucleosides commonly found in DNA and RNA, the property to form stable and extensive self-associations is limited to guanosine due to its unique hydrogen bonding donor and acceptor sites. Cations play a critical role in stabilizing G-quadruplex structures by occupying the central cavity and neutralizing the electrostatic repulsion of inwardly pointing guanine O6 oxygens. It was recognized early on that the ability to stabilize guanosine gels differed greatly between cations [9], suggesting that the ionic radius is important for complex stability. In the alkali series K+ promotes the most stable G-quadruplexes, followed by Rb+, Na+, Cs+, and Li+. Electrostatic effects are also likely to affect the relative ability of cations to stabilize G-quadruplexes [10]. The hydration energy of monovalent cations is inversely proportional to their ionic radii; hence the larger the cation the less hydrophilic it is, making it more likely to preferentially partition itself at the interior of the G-quartet. The same effect of different monovalent cations was also observed for the stability of structures formed by telomeric oligonucleotides, demonstrating that single-stranded telomeric DNA can fold into G-quadruplex structures under conditions within the physiological range [8].
Structural Diversity
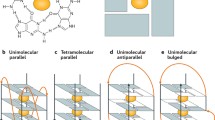
From the earliest days of studying G-quadruplexes in vitro, extensive structural polymorphism was noted. G-quadruplexes can be classified based on the participation of one (intramolecular) or two or more (intermolecular) DNA strands. DNA strands may be oriented in anti-parallel (Fig. 2a), parallel (Fig. 2b), or hybrid (Fig. 2c) configuration. Correspondingly, the nucleotide linkers between G-quartet stacks can adopt a multitude of loop structures (Fig. 2). G-quadruplex conformation is influenced by both the DNA sequence and the conditions used in the folding reaction such as the nature of the stabilizing cation. Although some general trends are apparent (e.g., potassium can favor parallel conformations [11]), there are always exceptions to these rules (e.g., both antiparallel potassium-stabilized and parallel sodium-stabilized G-quadruplexes exist and can be quite stable [12–15]). Thus it is difficult to predict the propensity of a sequence to fold into a particular structure, and each sequence needs to be characterized empirically under different folding conditions. The existence of multiple G-quadruplex conformations in equilibrium in the same solution [12, 16, 17] emphasizes the (often-overlooked) need to purify individual isomers prior to analysis [13, 18].
Human telomeric intramolecular G-quadruplexes. a Topology (i) and NMR structure (ii) of oligonucleotide AGGG(TTAGGG)3 in sodium containing solution, demonstrating an antiparallel conformation [134]. b Topology (i) and crystal structure (ii, iii) of oligonucleotide AGGG(TTAGGG)3 in potassium containing solution, showing a parallel “propeller” structure [27]. The crystal structure is shown as a side view (ii) and a top view (iii). c Hybrid conformations in potassium solution. Hybrid 1 (i) and hybrid 2 (ii) topologies illustrate differences in loop structures [32, 33]. The NMR structure of hybrid 2 is shown in (iii) [32]. Reproduced by permission of The Royal Society of Chemistry [138]
The stability of G-quadruplexes also varies widely; it depends not only on the identity of the stabilizing cation, but also on the DNA length and sequence, the length of intervening loops, and the strand stoichiometry and alignment [19–21]. There has been some recent progress in developing computational methods for predicting G-quadruplex stability, which will likely improve further as increasing amounts of empirical data are incorporated [22]. As a G-quartet contains eight hydrogen bonds in comparison to the two or three present in Watson–Crick base pairs, it might be expected that G-quadruplexes have equal or higher stability than duplex DNA. This is indeed often the case: many G-quadruplexes have melting temperatures well in excess of 60 or 70°C under otherwise physiological conditions [19]. This suggests that G-quadruplex DNA can potentially compete with duplex formation in vivo. In agreement, the molecular crowding agent polyethylene glycol, typically used to simulate the molecularly crowded intracellular environment, was demonstrated to favor formation of G-quadruplexes over duplex DNA [23, 24].
A good example of the heterogeneity of G-quadruplex structures is the intramolecular quadruplex formed from human telomeric sequence, which is of intense interest due to its ability to block telomere elongation by the cancer-associated enzyme telomerase in vitro [25]. The structures of no fewer than five different conformations of a human telomeric oligonucleotide containing four clusters of GGG have been solved to date. The NMR solution structure of the oligonucleotide AGGG(TTAGGG)3 in the presence of sodium is an anti-parallel basket-type quadruplex [26] (Fig. 2a), while the crystal structure of this oligonucleotide in the presence of potassium represents a parallel propeller-type intramolecular G-quadruplex [27] (Fig. 2b). Two different but related conformations have been detected in potassium solution, known as “hybrid” forms since they have both some parallel and antiparallel strands [28–31]. The solution structures of the two forms (“hybrid 1”, Fig. 2c(i) and “hybrid 2”, Fig. 2c(ii,iii)) reveal an identical G-quadruplex core structure with differences in the connecting loops [32, 33]. Recently, an antiparallel, basket-type human intramolecular G-quadruplex has also been demonstrated in potassium solution; this conformation is more stable than the “hybrid” forms despite the presence of only two G-quartet layers in the core [34]. The equilibrium between at least some of these potassium-stabilized conformations is influenced by the flanking sequences of the oligonucleotide, outside the GGG(TTAGGG)3 core. The presence of additional nucleotides at the 5′ end favors the hybrid over antiparallel conformations [35], while additional nucleotides at the 3′ end favor formation of hybrid 2 [32, 33].
Which of these structures is the physiologically relevant conformation has been the subject of much debate. The presence of 40% polyethylene glycol induced a shift from hybrid conformations to apparently parallel structures [36, 37]; however, the same effect was observed using acetonitrile, and is likely to be a result of dehydration rather than molecular crowding [38]. It is possible that all conformations are present in vivo under different conditions. In support of this, antiparallel, hybrid, and parallel conformations have all been observed to form from the same oligonucleotide in vitro at different concentrations of DNA [35]. The in vivo equilibrium may also be affected by temperature, ionic conditions, and the binding of particular proteins.
Recent studies have attempted to mimic the in vivo situation more closely by using longer telomeric oligonucleotides, since human telomeres end in a single-stranded 3′ G-rich overhang of 150–250 nucleotides [39–42]. Thermodynamic measurements support the existence of interconnecting intramolecular G-quadruplexes along a 72 nt telomeric oligonucleotide [43], and such “bead-on-a-string” conformations have been observed by atomic force microscopy [44]. Computer modeling combined with biophysical measurements suggests that an 8-repeat telomeric oligonucleotide forms into linked hybrid 1 and hybrid 2 G-quadruplex conformations [45]. Validation of these G-quadruplex conformations in vivo will require the development of conformation-specific probes suitable for use in human cells.
Biological Roles for G-Quadruplexes
As many nucleic acid sequences rich in guanosines are capable of forming G-quadruplexes, one wonders how prevalent these structures truly are within cells. Telomeric DNA has received much attention in this regard, in part because chromosomes end in single-stranded overhangs of the G-rich strand which may fold into G-quadruplex structures. But extended single-strandedness is not a prerequisite for G-quadruplex formation. Transient destabilization of duplex DNA during transcription, replication or DNA repair may well be sufficient to allow G-quadruplex DNA formation at many sites in the genome. Bioinformatic analysis has identified 375,000 candidate sequences within the human genome that could form G-quadruplex structures [46, 47]. It is possible that not all of these sequences form stable quadruplexes under physiological conditions [48]. However, the non-random distribution of potentially G-quadruplex forming sequences across the genome as well as the non-random length and sequence of loop regions argues that natural selection may be at work. Coding sequences are underrepresented for the transcribed strand suggesting that G-quartet formation in mRNA may be detrimental [46]. Despite the underrepresentation of coding sequences, the frequency at which potentially G-quadruplex forming sequences are found within transcribed regions displays an intriguing correlation with gene function. They are frequently found in protooncogenes including c-MYC, VEGF, c-kit, HIF-1a, and BCL2, but are significantly underrepresented in tumor suppressor genes [49].
A role for G-quadruplexes in gene regulation seems likely as putative G-quadruplex forming sequences are also concentrated in promoter regions. Nearly half of all known genes in the human genome harbor such sequences within 1000 nucleotides upstream of the transcription start site [50]. Regions of the human genome that are both within promoters and hypersensitive to nuclease cleavage show the greatest enrichment of potential quadruplex elements. The nuclease sensitivity of these sites indicates that the DNA is not bound by nucleosomes or other proteins and therefore may be more prone to G-quadruplex formation. This bias may at least in part reflect the G-richness of many transcription factor binding sites, such as Sp1 [51]. Careful examination of individual promoter sequences will be required to dissect the contributions of the different pathways and structures in gene regulation. For several genes, it has been experimentally confirmed that the G-quadruplex forming region plays a critical role in regulating expression of the gene. A single point mutation which destabilizes the G-quadruplex found in the c-MYC promoter resulted in a threefold increase in basal transcriptional activity of this gene [52]. Conversely, a cationic porphyrin known to stabilize a G-quadruplex structure was able to suppress c-MYC activation. These results strongly argue for a regulatory role of this particular G-quadruplex as a repressor of c-MYC transcription. This role may be mediated by the protein nucleolin, which has been shown to bind to this G-quadruplex both in vitro and in HeLa cells. Overexpression of nucleolin reduced expression from a c-MYC promoter reporter plasmid [53]. Nucleolin has also been implicated in binding to a G-quadruplex in the promoter of the VEGF gene; existence of this G-quadruplex was demonstrated by footprinting both in vitro and in vivo [54]. ReqQ helicase family members, such as budding yeast Sgs1 and human WRN and BLM, display potent quadruplex unwinding activity [55–57]. Consistent with a role for G-quadruplex structures in gene regulation, loci that are upregulated in cells harboring defects in one of these helicases correlate well with those that have high probability of forming G-quadruplex structures [58, 59]. Several groups have reported the potential formation of multiple G-quadruplexes in the promoter of the catalytic subunit of telomerase, hTERT [60–62], but a link between these G-quadruplexes and modulation of hTERT transcription has not yet been demonstrated.
Despite much circumstantial evidence in favor of the existence of telomeric G-quadruplex structures in human cells as well as tantalizing hints at potential functions, the actual role(s) of these structures in vivo have remained enigmatic. Intermolecular G-quadruplexes could facilitate telomere–telomere associations; such interactions have been observed in the telomere-rich environment of the macronuclei of ciliated protozoa and there is evidence that they are mediated by G-quadruplexes [63, 64]. It was postulated over 20 years ago that intermolecular parallel G-quadruplexes may be involved in the alignment of sister chromatids during meiosis [6], but direct evidence for this intriguing possibility is lacking. The clustering of telomeres in a meiotic bouquet arrangement has been observed in almost all organisms [65] and the demonstration that G-quadruplexes are involved in bouquet formation would represent a major advance in understanding this ubiquitous structure. Intriguingly a component of the meiosis-specific synaptonemal complex in S. cerevisiae, Hop1, was reported to promote pairing of double-stranded DNA helices via G-quartet formation, implicating intermolecular G-quadruplexes as the vehicles of chromosomal synapsis during meiotic prophase [66]. Furthermore, deletion of a G-quadruplex specific nuclease, KEM1, blocks meiosis in yeast, consistent with the hypothesis that G-quadruplex DNA may be involved in homologous chromosome pairing [67].
G-quadruplexes may cap telomeres by sequestering the single-stranded overhangs and preventing inappropriate elongation by telomerase, nucleolytic degradation, and end-to-end fusion. Intramolecular antiparallel G-quadruplexes have indeed been shown to be resistant to telomerase elongation in vivo [13, 25, 68], although parallel intermolecular structures are extended by telomerase [13]. There is some evidence in support of a protective role for G-quadruplexes; incubation of duplex DNA with human cell extract elicited a DNA damage response which was alleviated by addition of a 3′ tail capable of forming a G-quadruplex [69]. However, the possibility that the protective function was mediated by a telomere-binding protein in the extract was not ruled out in this study.
There are several reasons to believe that G-quadruplexes have regulatory functions at the level of RNA that may be more prevalent than of those in DNA. Firstly, RNA is single-stranded, at least when first synthesized, and although extensive Watson–Crick base-pairing occurs in some RNAs, a substantial portion of most RNAs lacks extensive regions of complementary sequences able to form continuous double-stranded helices. Secondly, G-quadruplexes are even more stable in RNA than in DNA and once formed they are highly refractory to unfolding [70]. Roles for G-quadruplexes in RNA regulation, splicing, and processing are further supported by the enrichment of candidate sequences in 5′ UTRs [71], first introns [51], and near polyadenylation signals [72]. The presence of a G-quadruplex was first experimentally verified in the 5′ UTR of NRAS and a repressive effect on translation was documented [71]. Since then, G-quadruplexes in the 5′ UTRs of more than 10 genes, including BCL-2, TRF2, and the gene for estrogen receptor α, have been demonstrated to downregulate translation [73–78]. Interestingly, a G-quadruplex in an IRES element of the VEGF gene has been shown to positively regulate translation [79]. As nearly 3000 mRNAs have potentially G-quadruplex forming sequences in their 5′ UTR it is tempting to speculate that G-quadruplex structures may be widely used to control gene expression at the translational level.
The fragile X mental retardation protein (FMRP) associates with polysomes and is thought to regulate mRNA translation. In vitro selection for RNAs that are preferentially bound by FMRP identified RNA ligands which form intramolecular G-quartets indicating that G-quadruplex containing mRNAs may be the target of FMRP regulation [80]. Indeed, when FMRP-containing ribonucleoprotein complexes were immunoprecipitated from mouse brain, nearly 70% of the associated mRNAs contained sequences predicted to form G-quartet structures [81]. Such strong correlation argues for a role of this structure in identifying the class of RNAs regulated by FMRP.
Alternative splicing of a number of genes is affected by G-rich sequences in the pre-mRNA and regulatory roles in vivo have been proposed. One of the most interesting examples was discovered in the context of studying the effects of G-quadruplex stabilizing drugs on telomerase activity in cancer cells. Early in vitro experiments had shown that stabilizing G-quartet structures in single-stranded telomeric DNA could inhibit elongation by telomerase [68]. It was therefore believed that the G-quartet stabilizing compound 12459 caused telomere shortening and apoptosis in a lung adenocarcinoma cell line by binding to the ends of chromosomes and inhibiting telomerase. However, closer examination revealed that the effect was largely mediated by stabilization of G-quartets in the pre-mRNA of the catalytic subunit of telomerase causing a shift in splicing pattern such that an inactive form of TERT is produced [82]. A G-quadruplex forming sequence in an intron of the tumor suppressor gene p53 has also been shown to modulate a shift to a splice variant of p53 that lacks the main transactivation domain [83]. To what extent G-quartet structures are involved in regulating alternative splicing of TERT and other genes in the absence of stabilizing compounds is presently unclear, but the potential for modulating the expression of many genes at this level is attractive.
Another potentially G-quadruplex forming RNA is telomeric repeat-containing RNA (TERRA) [84]. It appears that the C-rich strand of telomeric DNA is actively transcribed from several promoters within subtelomeric DNA and the G-rich RNA product remains associated with telomeric chromatin. As the complementary RNA was not detected, one would expect TERRA to form quadruplex structures unless prevented from doing so by interactions with proteins or telomeric DNA. Telomeric RNA oligonucleotides have been observed to form G-quadruplexes in vitro, but structural studies using both NMR and crystallography have so far revealed only parallel conformations in both sodium and potassium, showing that such G-quadruplexes display considerably less structural heterogeneity than their DNA counterparts [85–87]. TERRA G-quadruplexes are also considerably more stable than their DNA counterparts, at least in potassium, due to the propensity of the 2′ hydroxyl groups to engage in an increased number of hydrogen bonds [85, 88, 89]. There is some evidence that TERRA molecules can form into G-quadruplexes in vivo; introduction of a telomeric RNA oligonucleotide into HeLa cells enabled detection of a compact structure at telomeres by fluorescence resonance energy transfer (FRET) [90]. The role of this structure in TERRA function remains to be determined.
Given the abundance of nucleic acid sequences that can form G-quadruplex structures and the evidence supporting their formation under physiological conditions, there is little doubt that such structures form in vivo. There is also accumulating evidence that numerous proteins interact with G-quadruplex DNA and in some cases promote their unfolding. An issue that has been far more difficult to resolve is whether there is a positive regulatory role for G-quadruplex DNA in biology. The presence of a specific nucleic acid structure is inherently difficult to verify in vivo. Intracellular transcription of G-rich DNA in E. coli has been shown to produce loops of the non-template strand containing G-quadruplex structures that are detectable by electron microscopy [91]. Arguably the most direct evidence for G-quadruplex DNA existing in cells is that antibodies raised against G-quadruplex DNA label the macronuclei of a ciliate [64, 92]. A concern often raised about such experiments is that the reagent used for detection may drive the equilibrium towards the folded form, thus creating the very structure it is designed to detect. Nevertheless, Lipps and colleagues have used such antibodies in an intriguing series of experiments aimed at dissecting telomere structure throughout the cell cycle. Their work has led to a model in which telomere end binding proteins TEBP α and β actively stabilize G-quadruplexes for most of the cell cycle. During S-phase, TEBP β is phosphorylated and dissociates from the telomere. At the same time telomerase is recruited and G-quadruplex structures are resolved making the chromosome ends available for extension by telomerase [93, 94].
Applications for G-Quadruplex Stabilizing or Disrupting Compounds
In 1991, Zahler and colleagues demonstrated for the first time that an intramolecular telomeric G-quadruplex could not be extended by Oxytricha telomerase in vitro [68]. Based on this finding, a substantial effort has been made to identify synthetic and natural compounds that lock telomeric DNA in a G-quadruplex conformation and thus impede telomere elongation in vivo. Given the requirement for telomere maintenance in the indefinite proliferation of cancer cells, such molecules are promising candidates as anti-cancer drugs. A large number of G-quadruplex-interacting ligands from many chemical classes have been described [95, 96]. Those ligands which have been conclusively demonstrated to inhibit telomerase in vitro include the 2,6-diamidoanthraquinone BSU-1051 [97], the perylene diimide PIPER [98], the porphyrin TMPyP4 [99], the trisubstituted acridine BRACO19 [100, 101], bisquinolinium compounds such as 360A, 307A and the PhenDC series [100, 102, 103], and the natural product telomestatin [100, 104].
Telomestatin is one of the most well-studied G-quadruplex ligands due to its ability to greatly stabilize G-quadruplexes and its high specificity for these structures. Telomestatin induces and specifically recognizes the human intramolecular [105] antiparallel [106] G-quadruplex conformation. Telomestatin initially appeared to be a very potent telomerase inhibitor in vitro with an EC50 value of 5 nM [104], although this is now known to be at least an order of magnitude greater [100]. Nevertheless, at relatively low doses (≤2 μM), telomestatin causes gradual telomere shortening and growth arrest or apoptosis in a large number of cancer cell lines [107–112], supporting its use as a telomerase inhibitor in vivo. It has recently become clear, however, that classical telomerase inhibition is only part of the telomeric mechanism of action of telomestatin and related drugs. Higher doses of telomestatin (≥5 μM) lead to proliferation defects within a time frame that is too short for the effects to be explained by telomere shortening [107, 110]. This effect is independent of the telomerase status of the cells, and is likely due to direct uncapping of the chromosome termini in tumor cells. There are now several lines of evidence to support the uncapping mechanism; namely, treatment with telomestatin has been shown to cause degradation of the telomeric 3′ G-overhang [107, 110, 113], rapid dissociation of the telomere capping proteins TRF2 and POT1 from telomeric termini [110, 113, 114], and an increase in DNA damage signaling at the telomeres [113]. Other G-quadruplex stabilizing ligands such as BRACO19 and the pentacyclic acridine RHPS4 also cause disruption of the protective telomere cap structure [115–118].
It was initially envisaged that telomerase inhibition by G-quadruplex stabilizers would be a very specific cancer therapy, due to the absence of active telomerase in most normal tissues. A general effect on telomere structure raises the worrying possibility of toxic effects on normal cells. Nevertheless, several of the aforementioned drugs show good selectivity for cancer cell lines over normal cells, for unknown reasons [110–112, 118]. This may be due to a different telomere cap structure in normal versus cancer cells, or the existence of intact checkpoint pathways; these possibilities remain to be explored. This raises the exciting possibility that G-quadruplex stabilizers will constitute a specific cancer therapy that has the capability of overcoming the time-lag required for telomere shortening to occur.
Other considerations when evaluating potential telomere-targeted drugs include their specificity for particular G-quadruplex conformations, given the large number of potential G-quadruplex forming sequences in the human genome. For example, the porphyrin TMPyP4 interacts with telomeric G-quadruplexes with a minimal degree of specificity over its interaction with a G-quadruplex in the promoter of the c-Myc oncogene [119, 120]. The cellular effects of other ligands, however, are clearly mediated primarily through the telomeres; for example, overexpression of telomere proteins TRF2 and POT1 rendered xenograft tumors resistant to the effects of RHPS4 [118]. Furthermore, the implications of the extension of some types of G-quadruplexes by telomerase are also unknown [13]. While telomere-targeted G-quadruplex stabilizing molecules are showing great promise as anti-cancer drugs, their mechanisms of cellular action and the likelihood of adverse effects on healthy, proliferating cells must be further investigated prior to clinical use.
Methodologies Used to Study G-Quadruplex Structures
There are many simple techniques that can be used to probe aspects of the structure of a G-quadruplex. Native gel electrophoresis revealed early on that G-rich oligonucleotides have an unusual structure that results in aberrant migration on a non-denaturing acrylamide gel [5, 6], and this remains an accessible and straightforward technique to reveal the presence of a G-quadruplex. Intramolecular G-quadruplexes have a compact structure and thus migrate faster through a cation-containing gel than their linear counterparts [8], while intermolecular G-quadruplexes migrate slower due to increased molecular weight [6, 7]. Native gel electrophoresis is also invaluable in enabling purification of G-quadruplexes, an important consideration given the heterogeneity of structures that can form from a single oligonucleotide [121].
Other techniques are required to verify that the aberrantly migrating structures contain G-quartets. Circular dichroism (CD) spectroscopy is a convenient diagnostic tool in this regard, and has the additional advantage of being able to discriminate between G-quadruplex conformations. In this technique, the sample is exposed to circularly polarized light; if there is a chiral species in the solution, it will generally interact asymmetrically with the light, with the asymmetry varying with wavelength. Although it is difficult to predict a CD spectrum from a structure, characteristic spectra corresponding to different G-quadruplex conformations have been determined empirically. Parallel-stranded G-quadruplexes show a peak at 260 nm and a trough at 240 nm, while a peak at 295 nm and a trough at 260 nm are diagnostic of anti-parallel structures [122, 123]. The recently described “hybrid” structures formed from human telomeric oligonucleotides (Fig. 2c) show a strong peak at 290 nm with a shoulder out to about 270 nm, and troughs at 235 and 255 nm [28, 31].
Caution should be exercised when interpreting CD spectra, however; not only will unpurified mixtures of G-quadruplexes display spectra that may not be representative of their constituent conformations [18], but there are also exceptions to the above generalizations for characteristic spectra. The antiparallel human telomeric G-quadruplex in potassium solution has a CD spectrum very similar to that of a hybrid form, for example [34, 35], and a peak at 260 nm with a trough at 240 nm can also be displayed by non-quadruplex conformations such as duplexes, hairpins and single-stranded DNA [124]. Another type of spectrum that can provide a signature for nucleic acid structure is the UV thermal difference spectrum (TDS), i.e., the difference in UV spectra between the folded and unfolded states of a nucleic acid [125]. G-quadruplexes have a distinctive TDS [125], so combining data from a CD spectrum with a TDS increases the likelihood of correctly identifying a G-quadruplex.
If carried out over a range of temperatures, CD spectroscopy can also be used to observe melting of a G-quadruplex and hence determine thermodynamic parameters such as T m, ΔH, and ΔG 0, vital information for comparing stabilities of structures [12, 122, 126]. G-quadruplexes also show changes in UV absorbance at 295 nm relative to their linear counterparts, so UV spectroscopy may also be used to derive thermodynamic parameters that are reflective of G-quadruplex stability [126].
One of the earliest techniques used to verify G-quartet models of telomeric structure was dimethylsulfate (DMS) footprinting [6–8, 127]. DMS methylates the N7 position of guanine; subsequent treatment with piperidine breaks the DNA backbone at methylated sites. Gel electrophoresis allows visualization of the length of the cleaved fragments. In a G-quadruplex the N7 is hydrogen bonded and protected from methylation (see Fig. 1), resulting in little or no cleavage at the guanines involved in G-quartets [128].
A powerful recent method for probing G-quadruplex conformation and dynamics is single-molecule FRET [129]. In FRET, the oligonucleotide to be folded into a G-quadruplex is labeled with a donor and an acceptor fluorophore. Upon folding of the DNA, the donor fluorophore transfers its energy to the acceptor, with an efficiency that depends on their distance apart and relative orientation. By performing FRET on a dilute solution or a surface-immobilized sample, and capturing the resulting energy emission with a confocal fluorescence microscope, the dynamics of folding of a single-molecule can be observed; this removes the need to average the signal from a population of non-synchronously folding molecules, allowing sensitive dynamic analysis [130]. Application of this technique to the human intramolecular telomeric G-quadruplex has revealed that two conformations coexist in solution in both sodium and potassium buffers, and each conformation can be further divided into long-lived (minutes) and short-lived (seconds) species [16, 17].
The above methods provide a wealth of information about G-quadruplex behavior and conformation, but in order to determine precise molecular structures high-resolution techniques such as nuclear magnetic resonance (NMR) and X-ray crystallography are required. NMR first revealed the strand orientations and loop configurations of several telomeric G-quadruplexes and led to high-resolution structures [26, 131–135]. X-ray crystallography was also successful in generating high-resolution structures [136, 137]; there are now more than 30 reported structures of G-quadruplexes, some with resolutions less than 1 Å. In some cases, structures of the same molecule solved using both techniques differ either subtly [132, 136] or quite dramatically [27, 134]. It is likely that this is a result of the molecular crowding conditions introduced by crystallization. Which technique better represents the in vivo situation is equivocal, and further advances in technology will be needed to determine the true structure of G-quadruplexes within living cells.
References
Bang, I. (1910). Untersuchungen über die Guanylsäure. Biochemische Zeitschrift, 26, 293–311.
Gellert, M., Lipsett, M. N., & Davies, D. R. (1962). Helix formation by guanylic acid. Proceedings of National Academy of Science of the United States of America, 48, 2013–2018.
Chantot, J. F., & Guschlbauer, W. (1972). Mechanism of gel formation by guanine nucleosides. Jerusalem Symposium on Quantum Chemistry and Biochemistry, 4, 205–214.
Ralph, R. K., Connors, W. J., & Khorana, H. G. (1962). Secondary structure and aggregation in deoxyguanosine oligonucleotides. Journal of the American Chemical Society, 84, 2265–2266.
Henderson, E., Hardin, C. C., Walk, S. K., Tinoco, I., Jr., & Blackburn, E. H. (1987). Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell, 51, 899–908.
Sen, D., & Gilbert, W. (1988). Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature, 334, 364–366.
Sundquist, W. I., & Klug, A. (1989). Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature, 342, 825–829.
Williamson, J. R., Raghuraman, M. K., & Cech, T. R. (1989). Monovalent cation-induced structure of telomeric DNA: The G-quartet model. Cell, 59, 871–880.
Chantot, J., & Guschlbauer, W. (1969). Physicochemical properties of nucleosides 3. Gel formation by 8-bromoguanosine. FEBS Letter, 4, 173–176.
Williamson, J. R. (1994). G-quartet structures in telomeric DNA. Annual Review of Biophysics and Biomolecular Structure, 23, 703–730.
Miura, T., Benevides, J. M., & Thomas, G. J., Jr. (1995). A phase diagram for sodium and potassium ion control of polymorphism in telomeric DNA. Journal of Molecular Biology, 248, 233–238.
Balagurumoorthy, P., & Brahmachari, S. K. (1994). Structure and stability of human telomeric sequence. The Journal of Biological Chemistry, 269, 21858–21869.
Oganesian, L., Moon, I. K., Bryan, T. M., & Jarstfer, M. B. (2006). Extension of G-quadruplex DNA by ciliate telomerase. EMBO Journal, 25, 1148–1159.
Phan, A. T., & Patel, D. J. (2003). Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: Distinct topologies, thermodynamic properties, and folding/unfolding kinetics. Journal of the American Chemical Society, 125, 15021–15027.
Schultze, P., Smith, F. W., & Feigon, J. (1994). Refined solution structure of the dimeric quadruplex formed from the Oxytricha telomeric oligonucleotide d(GGGGTTTTGGGG). Structure, 2, 221–233.
Lee, J. Y., Okumus, B., Kim, D. S., & Ha, T. (2005). Extreme conformational diversity in human telomeric DNA. Proceedings of National Academy of Science of the United States of America, 102, 18938–18943.
Ying, L., Green, J. J., Li, H., Klenerman, D., & Balasubramanian, S. (2003). Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proceedings of National Academy of Science of the United States of America, 100, 14629–14634.
Dailey, M. M., Miller, M. C., Bates, P. J., Lane, A. N., & Trent, J. O. (2010). Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Research, 38, 4877–4888.
Hardin, C. C., Perry, A. G., & White, K. (2000). Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers, 56, 147–194.
Bugaut, A., & Balasubramanian, S. (2008). A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry, 47, 689–697.
Guedin, A., Gros, J., Alberti, P., & Mergny, J. L. (2010). How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Research, 38, 7858–7868.
Stegle, O., Payet, L., Mergny, J. L., MacKay, D. J., & Huppert, J. L. (2009). Predicting and understanding the stability of G-quadruplexes. Bioinformatics, 25, i374–i382.
Kan, Z. Y., Lin, Y., Wang, F., Zhuang, X. Y., Zhao, Y., Pang, D. W., et al. (2007). G-quadruplex formation in human telomeric (TTAGGG)4 sequence with complementary strand in close vicinity under molecularly crowded condition. Nucleic Acids Research, 35, 3646–3653.
Miyoshi, D., Matsumura, S., Nakano, S., & Sugimoto, N. (2004). Duplex dissociation of telomere DNAs induced by molecular crowding. Journal of the American Chemical Society, 126, 165–169.
Zaug, A. J., Podell, E. R., & Cech, T. R. (2005). Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proceedings of National Academy of Science of the United States of America, 102, 10864–10869.
Wang, Y., & Patel, D. J. (1993). Solution structure of a parallel-stranded G-quadruplex DNA. Journal of Molecular Biology, 234, 1171–1183.
Parkinson, G. N., Lee, M. P., & Neidle, S. (2002). Crystal structure of parallel quadruplexes from human telomeric DNA. Nature, 417, 876–880.
Ambrus, A., Chen, D., Dai, J., Bialis, T., Jones, R. A., & Yang, D. (2006). Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Research, 34, 2723–2735.
Luu, K. N., Phan, A. T., Kuryavyi, V., Lacroix, L., & Patel, D. J. (2006). Structure of the human telomere in K+ solution: An intramolecular (3 + 1) G-quadruplex scaffold. Journal of the American Chemical Society, 128, 9963–9970.
Phan, A. T., Luu, K. N., & Patel, D. J. (2006). Different loop arrangements of intramolecular human telomeric (3 + 1) G-quadruplexes in K+ solution. Nucleic Acids Research, 34, 5715–5719.
Xu, Y., Noguchi, Y., & Sugiyama, H. (2006). The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorganic and Medicinal Chemistry, 14, 5584–5591.
Dai, J., Carver, M., Punchihewa, C., Jones, R. A., & Yang, D. (2007). Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: Insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Research, 35, 4927–4940.
Phan, A. T., Kuryavyi, V., Luu, K. N., & Patel, D. J. (2007). Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Research, 35, 6517–6525.
Lim, K. W., Amrane, S., Bouaziz, S., Xu, W., Mu, Y., Patel, D. J., et al. (2009). Structure of the human telomere in K+ solution: A stable basket-type G-quadruplex with only two G-tetrad layers. Journal of the American Chemical Society, 131, 4301–4309.
Renciuk, D., Kejnovska, I., Skolakova, P., Bednarova, K., Motlova, J., & Vorlickova, M. (2009). Arrangements of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Research, 37, 6625–6634.
Xue, Y., Kan, Z. Y., Wang, Q., Yao, Y., Liu, J., Hao, Y. H., et al. (2007). Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. Journal of the American Chemical Society, 129, 11185–11191.
Zhou, J., Wei, C., Jia, G., Wang, X., Tang, Q., Feng, Z., et al. (2008). The structural transition and compaction of human telomeric G-quadruplex induced by excluded volume effect under cation-deficient conditions. Biophysical Chemistry, 136, 124–127.
Miller, M. C., Buscaglia, R., Chaires, J. B., Lane, A. N., & Trent, J. O. (2010). Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3′ sequence of d(AG 3(TTAG3)3). Journal of the American Chemical Society, 132, 17105–17107.
Chai, W., Du, Q., Shay, J. W., & Wright, W. E. (2006). Human telomeres have different overhang sizes at leading versus lagging strands. Molecular Cell, 21, 427–435.
Makarov, V. L., Hirose, Y., & Langmore, J. P. (1997). Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell, 88, 657–666.
McElligott, R., & Wellinger, R. J. (1997). The terminal DNA structure of mammalian chromosomes. EMBO Journal, 16, 3705–3714.
Wright, W. E., Tesmer, V. M., Huffman, K. E., Levene, S. D., & Shay, J. W. (1997). Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes and Development, 11, 2801–2809.
Yu, H. Q., Miyoshi, D., & Sugimoto, N. (2006). Characterization of structure and stability of long telomeric DNA G-quadruplexes. Journal of the American Chemical Society, 128, 15461–15468.
Xu, Y., Ishizuka, T., Kurabayashi, K., & Komiyama, M. (2009). Consecutive formation of G-quadruplexes in human telomeric-overhang DNA: A protective capping structure for telomere ends. Angewandte Chemie, 48, 7833–7836.
Petraccone, L., Trent, J. O., & Chaires, J. B. (2008). The tail of the telomere. Journal of the American Chemical Society, 130, 16530–16532.
Huppert, J. L., & Balasubramanian, S. (2005). Prevalence of quadruplexes in the human genome. Nucleic Acids Research, 33, 2908–2916.
Todd, A. K., Johnston, M., & Neidle, S. (2005). Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Research, 33, 2901–2907.
Risitano, A., & Fox, K. R. (2004). Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Research, 32, 2598–2606.
Eddy, J., & Maizels, N. (2006). Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Research, 34, 3887–3896.
Huppert, J. L., & Balasubramanian, S. (2007). G-quadruplexes in promoters throughout the human genome. Nucleic Acids Research, 35, 406–413.
Eddy, J., & Maizels, N. (2008). Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Research, 36, 1321–1333.
Siddiqui-Jain, A., Grand, C. L., Bearss, D. J., & Hurley, L. H. (2002). Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proceedings of National Academy of Science of the United States of America, 99, 11593–11598.
Gonzalez, V., Guo, K., Hurley, L., & Sun, D. (2009). Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. The Journal of Biological Chemistry, 284, 23622–23635.
Sun, D., Guo, K., & Shin, Y. J. (2010). Evidence of the formation of G-quadruplex structures in the promoter region of the human vascular endothelial growth factor gene. Nucleic Acids Research, 39, 1256–1265.
Fry, M., & Loeb, L. A. (1999). Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. The Journal of Biological Chemistry, 274, 12797–12802.
Huber, M. D., Lee, D. C., & Maizels, N. (2002). G4 DNA unwinding by BLM and Sgs1p: Substrate specificity and substrate-specific inhibition. Nucleic Acids Research, 30, 3954–3961.
Mohaghegh, P., Karow, J. K., Brosh, R. M., Jr, Bohr, V. A., Jr., & Hickson, I. D. (2001). The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Research, 29, 2843–2849.
Hershman, S. G., Chen, Q., Lee, J. Y., Kozak, M. L., Yue, P., Wang, L. S., et al. (2008). Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Research, 36, 144–156.
Johnson, J. E., Cao, K., Ryvkin, P., Wang, L. S., & Johnson, F. B. (2010). Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Research, 38, 1114–1122.
Lim, K. W., Lacroix, L., Yue, D. J., Lim, J. K., Lim, J. M., & Phan, A. T. (2010). Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. Journal of the American Chemical Society, 132, 12331–12342.
Micheli, E., Martufi, M., Cacchione, S., De Santis, P., & Savino, M. (2010). Self-organization of G-quadruplex structures in the hTERT core promoter stabilized by polyaminic side chain perylene derivatives. Biophysical Chemistry, 153, 43–53.
Palumbo, S. L., Ebbinghaus, S. W., & Hurley, L. H. (2009). Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. Journal of the American Chemical Society, 131, 10878–10891.
Lipps, H. J. (1980). In vitro aggregation of the gene-sized DNA molecules of the ciliate Stylonychia mytilus. Proceedings of National Academy of Science of the United States of America, 77, 4104–4107.
Schaffitzel, C., Berger, I., Postberg, J., Hanes, J., Lipps, H. J., & Pluckthun, A. (2001). In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proceedings of National Academy of Science of the United States of America, 98, 8572–8577.
Harper, L., Golubovskaya, I., & Cande, W. Z. (2004). A bouquet of chromosomes. Journal of Cell Science, 117, 4025–4032.
Anuradha, S., & Muniyappa, K. (2004). Meiosis-specific yeast Hop1 protein promotes synapsis of double-stranded DNA helices via the formation of guanine quartets. Nucleic Acids Research, 32, 2378–2385.
Liu, Z., & Gilbert, W. (1994). The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: Implication of in vivo functions for this novel DNA structure. Cell, 77, 1083–1092.
Zahler, A. M., Williamson, J. R., Cech, T. R., & Prescott, D. M. (1991). Inhibition of telomerase by G-quartet DNA structures. Nature, 350, 718–720.
Tsai, Y. C., Qi, H., & Liu, L. F. (2007). Protection of DNA ends by telomeric 3’ G-tail sequences. The Journal of Biological Chemistry, 282, 18786–18792.
Sacca, B., Lacroix, L., & Mergny, J. L. (2005). The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Research, 33, 1182–1192.
Kumari, S., Bugaut, A., Huppert, J. L., & Balasubramanian, S. (2007). An RNA G-quadruplex in the 5’ UTR of the NRAS proto-oncogene modulates translation. Nature Chemical Biology, 3, 218–221.
Kikin, O., Zappala, Z., D’Antonio, L., & Bagga, P. S. (2008). GRSDB2 and GRS_UTRdb: Databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Research, 36, D141–D148.
Arora, A., Dutkiewicz, M., Scaria, V., Hariharan, M., Maiti, S., & Kurreck, J. (2008). Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA, 14, 1290–1296.
Balkwill, G. D., Derecka, K., Garner, T. P., Hodgman, C., Flint, A. P., & Searle, M. S. (2009). Repression of translation of human estrogen receptor alpha by G-quadruplex formation. Biochemistry, 48, 11487–11495.
Beaudoin, J. D., & Perreault, J. P. (2010). 5’-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Research, 38, 7022–7036.
Gomez, D., Guedin, A., Mergny, J. L., Salles, B., Riou, J. F., Teulade-Fichou, M. P., et al. (2010). A G-quadruplex structure within the 5’-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Research, 38, 7187–7198.
Morris, M. J., & Basu, S. (2009). An unusually stable G-quadruplex within the 5’-UTR of the MT3 matrix metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry, 48, 5313–5319.
Shahid, R., Bugaut, A., & Balasubramanian, S. (2010). The BCL-2 5’ untranslated region contains an RNA G-quadruplex-forming motif that modulates protein expression. Biochemistry, 49, 8300–8306.
Morris, M. J., Negishi, Y., Pazsint, C., Schonhoft, J. D., & Basu, S. (2010). An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. Journal of the American Chemical Society, 132, 17831–17839.
Darnell, J. C., Jensen, K. B., Jin, P., Brown, V., Warren, S. T., & Darnell, R. B. (2001). Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell, 107, 489–499.
Brown, V., et al. (2001). Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell, 107, 477–487.
Gomez, D., Lemarteleur, T., Lacroix, L., Mailliet, P., Mergny, J. L., & Riou, J. F. (2004). Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Research, 32, 371–379.
Marcel, V. et al. (2010). G-quadruplex structures in TP53 intron 3: Role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis, 32, 271–278.
Azzalin, C. M., Reichenbach, P., Khoriauli, L., Giulotto, E., & Lingner, J. (2007). Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science, 318, 798–801.
Collie, G. W., Haider, S. M., Neidle, S., & Parkinson, G. N. (2010). A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Research, 38, 5569–5580.
Martadinata, H., & Phan, A. T. (2009). Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K+ solution. Journal of the American Chemical Society, 131, 2570–2579.
Xu, Y., Kaminaga, K., & Komiyama, M. (2008). G-quadruplex formation by human telomeric repeats-containing RNA in Na + solution. Journal of the American Chemical Society, 130, 11179–11184.
Arora, A., & Maiti, S. (2009). Differential biophysical behavior of human telomeric RNA and DNA quadruplex. Journal of Physical Chemistry B, 113, 10515–10520.
Joachimi, A., Benz, A., & Hartig, J. S. (2009). A comparison of DNA and RNA quadruplex structures and stabilities. Bioorganic and Medicinal Chemistry, 17, 6811–6815.
Xu, Y., Suzuki, Y., Ito, K., & Komiyama, M. (2010). Telomeric repeat-containing RNA structure in living cells. Proceedings of National Academy of Science of the United States of America, 107, 14579–14584.
Duquette, M. L., Handa, P., Vincent, J. A., Taylor, A. F., & Maizels, N. (2004). Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes and Development, 18, 1618–1629.
Schaffitzel, C., Postberg, J., Paeschke, K., & Lipps, H. J. (2010). Probing telomeric G-quadruplex DNA structures in cells with in vitro generated single-chain antibody fragments. Methods in Molecular Biology, 608, 159–181.
Paeschke, K., Juranek, S., Simonsson, T., Hempel, A., Rhodes, D., & Lipps, H. J. (2008). Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nature Structural and Molecular Biology, 15, 598–604.
Paeschke, K., Simonsson, T., Postberg, J., Rhodes, D., & Lipps, H. J. (2005). Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nature Structural and Molecular Biology, 12, 847–854.
De Cian, A., Lacroix, L., Douarre, C., Temime-Smaali, N., Trentesaux, C., Riou, J. F., et al. (2008). Targeting telomeres and telomerase. Biochimie, 90, 131–155.
Monchaud, D., & Teulade-Fichou, M. P. (2008). A hitchhiker’s guide to G-quadruplex ligands. Organic and Biomolecular Chemistry, 6, 627–636.
Sun, D., et al. (1997). Inhibition of human telomerase by a G-quadruplex-interactive compound. Journal of Medicinal Chemistry, 40, 2113–2116.
Fedoroff, O. Y., Salazar, M., Han, H., Chemeris, V. V., Kerwin, S. M., & Hurley, L. H. (1998). NMR-Based model of a telomerase-inhibiting compound bound to G-quadruplex DNA. Biochemistry, 37, 12367–12374.
Wheelhouse, R. T., Sun, D. K., Han, H. Y., Han, F. X., & Hurley, L. H. (1998). Cationic porphyrins as telomerase inhibitors: The interaction of tetra-(N-methyl-4-pyridyl)porphine with quadruplex DNA. Journal of the American Chemical Society, 120, 3261–3262.
De Cian, A., et al. (2007). Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proceedings of National Academy of Science of the United States of America, 104, 17347–17352.
Read, M., et al. (2001). Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proceedings of National Academy of Science of the United States of America, 98, 4844–4849.
De Cian, A., Delemos, E., Mergny, J. L., Teulade-Fichou, M. P., & Monchaud, D. (2007). Highly efficient G-quadruplex recognition by bisquinolinium compounds. Journal of the American Chemical Society, 129, 1856–1857.
Pennarun, G., et al. (2005). Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene, 24, 2917–2928.
Shin-ya, K., Wierzba, K., Matsuo, K., Ohtani, T., Yamada, Y., Furihata, K., et al. (2001). Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. Journal of the American Chemical Society, 123, 1262–1263.
Kim, M. Y., Vankayalapati, H., Shin-Ya, K., Wierzba, K., & Hurley, L. H. (2002). Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular g-quadruplex. Journal of the American Chemical Society, 124, 2098–2099.
Rezler, E. M., Seenisamy, J., Bashyam, S., Kim, M. Y., White, E., Wilson, W. D., et al. (2005). Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. Journal of the American Chemical Society, 127, 9439–9447.
Gomez, D., Paterski, R., Lemarteleur, T., Shin-Ya, K., Mergny, J. L., & Riou, J. F. (2004). Interaction of telomestatin with the telomeric single-strand overhang. The Journal of Biological Chemistry, 279, 41487–41494.
Kim, M. Y., Gleason-Guzman, M., Izbicka, E., Nishioka, D., & Hurley, L. H. (2003). The different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-quadruplex structures. Cancer Research, 63, 3247–3256.
Shammas, M. A., Shmookler Reis, R. J., Li, C., Koley, H., Hurley, L. H., Anderson, K. C., et al. (2004). Telomerase inhibition and cell growth arrest after telomestatin treatment in multiple myeloma. Clinical Cancer Research, 10, 770–776.
Tahara, H., Shin-Ya, K., Seimiya, H., Yamada, H., Tsuruo, T., & Ide, T. (2006). G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3’ telomeric overhang in cancer cells. Oncogene, 25, 1955–1966.
Tauchi, T., Shin-Ya, K., Sashida, G., Sumi, M., Nakajima, A., Shimamoto, T., et al. (2003). Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: Involvement of ATM-dependent DNA damage response pathways. Oncogene, 22, 5338–5347.
Tauchi, T., Shin-ya, K., Sashida, G., Sumi, M., Okabe, S., Ohyashiki, J. H., et al. (2006). Telomerase inhibition with a novel G-quadruplex-interactive agent, telomestatin: In vitro and in vivo studies in acute leukemia. Oncogene, 25, 5719–5725.
Gomez, D., et al. (2006). Telomestatin-induced telomere uncapping is modulated by POT1 through G-overhang extension in HT1080 human tumor cells. The Journal of Biological Chemistry, 281, 38721–38729.
Gomez, D., et al. (2006). The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Research, 66, 6908–6912.
Burger, A. M., Dai, F., Schultes, C. M., Reszka, A. P., Moore, M. J., Double, J. A., et al. (2005). The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Research, 65, 1489–1496.
Leonetti, C., et al. (2004). Biological activity of the G-quadruplex ligand RHPS4 (3, 11-difluoro-6, 8, 13-trimethyl-8H-quino[4, 3, 2-kl]acridinium methosulfate) is associated with telomere capping alteration. Molecular Pharmacology, 66, 1138–1146.
Phatak, P., Cookson, J. C., Dai, F., Smith, V., Gartenhaus, R. B., Stevens, M. F., et al. (2007). Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism. British Journal of Cancer, 96, 1223–1233.
Salvati, E., et al. (2007). Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. Journal of Clinical Investigation, 117, 3236–3247.
Halder, K., & Chowdhury, S. (2007). Quadruplex-coupled kinetics distinguishes ligand binding between G4 DNA motifs. Biochemistry, 46, 14762–14770.
Lemarteleur, T., Gomez, D., Paterski, R., Mandine, E., Mailliet, P., & Riou, J. F. (2004). Stabilization of the c-myc gene promoter quadruplex by specific ligands’ inhibitors of telomerase. Biochemical and Biophysical Research Communications, 323, 802–808.
Moon, I. K., & Jarstfer, M. B. (2010). Preparation of G-quartet structures and detection by native gel electrophoresis. Methods in Molecular Biology, 608, 51–63.
Balagurumoorthy, P., Brahmachari, S. K., Mohanty, D., Bansal, M., & Sasisekharan, V. (1992). Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Research, 20, 4061–4067.
Hardin, C. C., Henderson, E., Watson, T., & Prosser, J. K. (1991). Monovalent cation induced structural transitions in telomeric DNAs: G-DNA folding intermediates. Biochemistry, 30, 4460–4472.
Kypr, J., Kejnovska, I., Renciuk, D., & Vorlickova, M. (2009). Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Research, 37, 1713–1725.
Mergny, J. L., Li, J., Lacroix, L., Amrane, S., & Chaires, J. B. (2005). Thermal difference spectra: A specific signature for nucleic acid structures. Nucleic Acids Research, 33, e138.
Olsen, C. M., & Marky, L. A. (2010). Monitoring the temperature unfolding of G-quadruplexes by UV and circular dichroism spectroscopies and calorimetry techniques. Methods in Molecular Biology, 608, 147–158.
Panyutin, I. G., Kovalsky, O. I., Budowsky, E. I., Dickerson, R. E., Rikhirev, M. E., & Lipanov, A. A. (1990). G-DNA: A twice-folded DNA structure adopted by single-stranded oligo(dG) and its implications for telomeres. Proceedings of National Academy of Science of the United States of America, 87, 867–870.
Sun, D., & Hurley, L. H. (2010). Biochemical techniques for the characterization of G-quadruplex structures: EMSA, DMS footprinting, and DNA polymerase stop assay. Methods in Molecular Biology, 608, 65–79.
Okumus, B., & Ha, T. (2010). Real-time observation of G-quadruplex dynamics using single-molecule FRET microscopy. Methods in Molecular Biology, 608, 81–96.
Ha, T., Enderle, T., Ogletree, D. F., Chemla, D. S., Selvin, P. R., & Weiss, S. (1996). Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proceedings of National Academy of Science of the United States of America, 93, 6264–6268.
Smith, F. W., & Feigon, J. (1992). Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature, 356, 164–168.
Smith, F. W., & Feigon, J. (1993). Strand orientation in the DNA quadruplex formed from the Oxytricha telomere repeat oligonucleotide d(G4T4G4) in solution. Biochemistry, 32, 8682–8692.
Wang, Y., & Patel, D. J. (1992). Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplexes with anti glycosidic torsion angles in solution. Biochemistry, 31, 8112–8119.
Wang, Y., & Patel, D. J. (1993). Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure, 1, 263–282.
Wang, Y., & Patel, D. J. (1994). Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure, 2, 1141–1156.
Kang, C., Zhang, X., Ratliff, R., Moyzis, R., & Rich, A. (1992). Crystal structure of four-stranded Oxytricha telomeric DNA. Nature, 356, 126–131.
Laughlan, G., Murchie, A. I., Norman, D. G., Moore, M. H., Moody, P. C., Lilley, D. M., et al. (1994). The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science, 265, 520–524.
Bryan, T. (Ed.). (2009). Molecular Themes in DNA Replication (p. 264). Cambridge: Lynne Cox, Royal Society of Chemistry.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Bryan, T.M., Baumann, P. G-Quadruplexes: From Guanine Gels to Chemotherapeutics. Mol Biotechnol 49, 198–208 (2011). https://doi.org/10.1007/s12033-011-9395-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-011-9395-5