Abstract
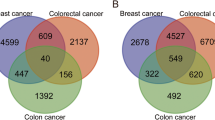
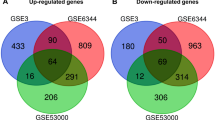
Metastasis of clear cell renal cell carcinoma (ccRCC) is a leading cause of death. The purpose of this research was to investigate the key gene in ccRCC tumor metastasis. Three microarray datasets (GSE22541, GSE85258, and GSE105261), which included primary and metastatic ccRCC tissues, were obtained from the Gene Expression Omnibus (GEO) database. Expression profiling and clinical data of ccRCC were downloaded from The Cancer Genome Atlas (TCGA) dataset. A total of 20 overlapping differentially expressed genes (DEGs) were identified using the R limma package. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis indicated that the DEGs were mainly enriched in tumor metastasis-related pathways. Gene expression analysis and survival analysis in the GEPIA2 database further identified the key gene HSD11B2. qRT-PCR result manifested that HSD11B2 level was significantly down-regulated in ccRCC tissues compared with adjacent normal tissues. ROC analysis showed that HSD11B2 exhibited good diagnostic efficiency for metastatic and non-metastatic ccRCC. Univariate and multivariate Cox regression analysis showed that HSD11B2 expression was an independent prognostic factor. To establish a nomogram combining HSD11B2 expression and clinical factors, and a new method for predicting the survival probability of ccRCC patients. Gene Set Enrichment Analysis (GSEA) enrichment results showed that low expression of HSD11B2 was mainly enriched in tumor signaling pathways and immune-related pathways. Immune analysis revealed a significant correlation between HSD11B2 and tumor immune infiltrates in ccRCC. This study suggests that HSD11B2 can serve as a potential biomarker and therapeutic target for ccRCC metastasis.






Similar content being viewed by others
Data availability
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article. All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Abbreviations
- ccRCC:
-
Clear cell renal cell carcinoma
- RCC:
-
Renal cell carcinoma
- DEGs:
-
Differentially expressed genes
- GEO:
-
Gene expression omnibus
- GO:
-
Gene ontology
- OS:
-
Overall survival
- TCGA:
-
The cancer genome atlas
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- qRT-PCR:
-
Quantitative real-time PCR
- GSEA:
-
Gene set enrichment analysis
- GEPIA2:
-
The gene expression profiling interactive analysis 2
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics. CA: Cancer J Clin. 2021;71(1):7–33.
Padala SA, Barsouk A, Thandra KC, Saginala K, Mohammed A, Vakiti A, Rawla P, Barsouk A. Epidemiology of renal cell carcinoma. World J Oncol. 2020;11(3):79–87.
Linehan WM, Ricketts CJ. The cancer genome atlas of renal cell carcinoma: findings and clinical implications. Nat Rev Urol. 2019;16(9):539–52.
Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009.
Gao S, Yan L, Zhang H, Fan X, Jiao X, Shao F. Identification of a metastasis-associated gene signature of clear cell renal cell carcinoma. Front Genet. 2020;11:603455.
Lalani AKA, McGregor BA, Albiges L, Choueiri TK, Motzer R, Powles T, Wood C, Bex A. Systemic treatment of metastatic clear cell renal cell carcinoma in 2018: current paradigms, use of immunotherapy, and future directions. Eur Urol. 2019;75(1):100–10.
Sun M, Lughezzani G, Perrotte P, Karakiewicz PI. Treatment of metastatic renal cell carcinoma. Nat Rev Urol. 2010;7(6):327–38.
Lopez JI, Cortes JM. Multisite tumor sampling: a new tumor selection method to enhance intratumor heterogeneity detection. Hum Pathol. 2017;64:1–6.
Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–9.
Kapitsinou PP, Haase VH. The VHL tumor suppressor and HIF: insights from genetic studies in mice. Cell Death Differ. 2008;15(4):650–9.
Dizman N, Philip EJ, Pal SK. Genomic profiling in renal cell carcinoma. Nat Rev Nephrol. 2020;16(8):435–51.
Ni L, Yuan C, Zhang C, Xiang Y, Wu J, Wang X, Wu X. Co-expression network analysis identified LTF in association with metastasis risk and prognosis in clear cell renal cell carcinoma. Onco Targets Ther. 2020;13:6975–86.
Sun Y, Zou J, Ouyang W, Chen K. Identification of as a potential core gene involved in the metastasis of clear cell renal cell carcinoma. Cancer Manage Res. 2020;12:5701–12.
Ho TH, Serie DJ, Parasramka M, Cheville JC, Bot BM, Tan W, Wang L, Joseph RW, Hilton T, Leibovich BC, et al. Differential gene expression profiling of matched primary renal cell carcinoma and metastases reveals upregulation of extracellular matrix genes. Ann Oncol. 2017;28(3):604–10.
Nam HY, Chandrashekar DS, Kundu A, Shelar S, Kho EY, Sonpavde G, Naik G, Ghatalia P, Livi CB, Varambally S, et al. Integrative epigenetic and gene expression analysis of renal tumor progression to metastasis. Mol Cancer Res. 2019;17(1):84–96.
Wuttig D, Zastrow S, Fussel S, Toma MI, Meinhardt M, Kalman K, Junker K, Sanjmyatav J, Boll K, Hackermuller J, et al. CD31, EDNRB and TSPAN7 are promising prognostic markers in clear-cell renal cell carcinoma revealed by genome-wide expression analyses of primary tumors and metastases. Int J Cancer. 2012;131(5):E693-704.
Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7.
Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–60.
Zhang Z, Lin E, Zhuang H, Xie L, Feng X, Liu J, Yu Y. Construction of a novel gene-based model for prognosis prediction of clear cell renal cell carcinoma. Cancer Cell Int. 2020;20:27.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50.
Salgia NJ, Dara Y, Bergerot P, Salgia M, Pal SK. The changing landscape of management of metastatic renal cell carcinoma: current treatment options and future directions. Curr Treat Options Oncol. 2019;20(5):41.
Lopez-Fernandez E, Lopez JI. The impact of tumor eco-evolution in renal cell carcinoma sampling. Cancers (Basel). 2018;10(12):485.
Mouw JK, Ou G, Weaver VM. Extracellular matrix assembly: a multiscale deconstruction. Nat Rev Mol Cell Biol. 2014;15(12):771–85.
Kai F, Drain AP, Weaver VM. The extracellular matrix modulates the metastatic journey. Dev Cell. 2019;49(3):332–46.
Northey JJ, Przybyla L, Weaver VM. Tissue force programs cell fate and tumor aggression. Cancer Discov. 2017;7(11):1224–37.
Lucotti S, Muschel RJ. Platelets and metastasis: new implications of an old interplay. Front Oncol. 2020;10:1350.
Chapman K, Holmes M, Seckl J. 11beta-hydroxysteroid dehydrogenases: intracellular gate-keepers of tissue glucocorticoid action. Physiol Rev. 2013;93(3):1139–206.
Agarwal AK, Rogerson FM, Mune T, White PC. Gene structure and chromosomal localization of the human HSD11K gene encoding the kidney (type 2) isozyme of 11 beta-hydroxysteroid dehydrogenase. Genomics. 1995;29(1):195–9.
Zhang MZ, Xu J, Yao B, Yin H, Cai Q, Shrubsole MJ, Chen X, Kon V, Zheng W, Pozzi A, et al. Inhibition of 11beta-hydroxysteroid dehydrogenase type II selectively blocks the tumor COX-2 pathway and suppresses colon carcinogenesis in mice and humans. J Clin Invest. 2009;119(4):876–85.
Qi L, Zhang Y, Song F, Ding Y. Chinese herbal medicine promote tissue differentiation in colorectal cancer by activating HSD11B2. Arch Biochem Biophys. 2020;695:108644.
Acknowledgements
Thanks to contributors to the GEO database and TCGA database for their platforms and datasets. And thanks to the researchers and study participants for their contributions.
Funding
This work was supported by a grant from The Intelligence Medicine Project of Chongqing Medical University, China (No. ZHYX2019015 to Longke Ran).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. LR and HW designed the experimental flow. SM collected and downloaded the data. JS and QL analyzed the data. JL and SM wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declared no conflict of interest.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. The study was approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (The ethical approval number: 2021-465). In addition, A preprint has previously been published in Research Square (link:https://www.researchsquare.com/article/rs-885592/v1). This is a preprint, a preliminary version of a manuscript that has not completed peer review at a journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Miao, S., Song, J., Liu, Q. et al. Integrated bioinformatics analysis to identify the key gene associated with metastatic clear cell renal cell carcinoma. Med Oncol 39, 128 (2022). https://doi.org/10.1007/s12032-022-01706-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-022-01706-y