Abstract
Lipid metabolism reprogramming is one of the adaptive events that drive tumor development and survival, and may account for resistance to chemotherapeutic drugs. Perilipins are structural proteins associated with lipophagy and lipid droplet integrity, and their overexpression is associated with tumor aggressiveness. Here, we sought to explore the role of lipid droplet-related protein perilipin-3 (PLIN3) in prostate cancer (PCa) chemotherapy. We investigated the role of PLIN3 suppression in docetaxel cytotoxic activity in PCa cell lines. Additional effects of PLIN3 depletion on autophagy-related proteins and gene expression patterns, apoptotic potential, proliferation rate, and ATP levels were examined. Depletion of PLIN3 resulted in docetaxel resistance, accompanied by enhanced autophagic flux. We further assessed the synergistic effect of autophagy suppression with chloroquine on docetaxel cytotoxicity. Inhibition of autophagy with chloroquine reversed chemoresistance of stably transfected shPLIN3 PCa cell lines, with no effect on the parental ones. The shPLIN3 cell lines also exhibited reduced Caspase-9 related apoptosis initiation. Moreover, we assessed PLIN3 expression in a series of PCa tissue specimens, were complete or partial loss of PLIN3 expression was frequently noted in 70% of the evaluated specimens. Following PLIN3 silencing, PCa cells were characterized by impaired lipophagy and acquired an enhanced autophagic response upon docetaxel-induced cytotoxic stress. Such an adaptation leads to resistance to docetaxel, which could be reversed by the autophagy blocker chloroquine. Given the frequent loss of PLIN3 expression in PCa specimens, we suggest that combination of docetaxel with chloroquine may improve the efficacy of docetaxel treatment in PLIN3-deficient cancer patients.
Similar content being viewed by others

Introduction
Prostate Cancer (PCa) is the second most diagnosed malignancy and the fifth most common cause of cancer-related deaths among men worldwide, counting roughly 1.3 million new cases and 360,000 deaths in 2018 [1]. Radical prostatectomy and radiotherapy offer high cure rates in early stages of the disease, whereas for metastatic hormone-sensitive PCa, androgen deprivation therapy (ADT) is the treatment of choice [2, 3]. Even though patients respond well to ADT, they usually develop advanced metastatic castration-resistant PCa (mCRPCa) [4] and, then, chemotherapy is added to the standard treatment. For patients with metastatic castration-resistant PCa, docetaxel chemotherapy offers a survival benefit [5,6,7,8]. Docetaxel is an antineoplastic drug and one of the few drugs approved for treatment of patients with metastatic PCa. Docetaxel binds and stabilizes cellular microtubule filaments, leading to proliferation inhibition, impaired mitotic spindle formation and cell death via apoptosis and mitotic catastrophe [9].
Numerous factors steer cancer resistance to cytotoxic stimuli induced by chemotherapeutic agents, resulting in tumor relapse and disease progression [10]. Metabolic reprogramming is one of them and also considered an emerging cancer hallmark [11]. During cancer progression, tumors tend to alter or enhance their lipid metabolism, as a consequence of the reduced nutrient and oxygen supply. This alteration facilitates their adaptation to their unique microenvironment [12, 13]. Interestingly, lipophagy is also involved in this metabolic shift [14]. Lipophagy is a recently introduced term that describes the close interplay between the autophagy pathway and Lipid Droplets’ (LDs) breakdown. Furthermore, the majority of autophagy-related proteins are also involved in lipophagy [15]. Lipophagy is a regulatory process, as cells can utilize different amount of lipids, according to their needs; these molecules are sequestered for degradation by the autophagic machinery to provide the essential energy through β-oxidation [15, 16]. In dividing cells, intracellular lipids are metabolized in order to generate ATP, sustain redox balance, provide necessary components for new membranes, and regulate cell signaling processes [16,17,18]. Therefore, lipophagy is an essential and elaborately regulated cellular function, responsible for intracellular degradation of accumulated LDs, maintenance of intracellular levels of fatty acids (FAs), and collectively, for cellular energy homeostasis [17, 18].
LDs are essential and dynamic organelles that function as storage of intracellular neutral lipids, mainly triglycerides and cholesterol [19, 20]. LDs consist of a hydrophobic core -where neutral lipids are actually stored- surrounded by a phospholipid monolayer membrane. LD-associated proteins, known as PAT proteins (named after Perilipin/ADRP/TIP47), are found on the LD surface [21]. PAT proteins belong to a recently discovered family of five proteins associated with LDs, known as perilipins. Members of this family are PLIN1 (Perilipin), PLIN2 (ADRP or Adipophilin), PLIN3 (TIP47), PLIN4 (S3-12), and PLIN5 (OXPAT). Perilipins are considered essential for the development, integrity, and modification of LDs [21, 22]. Moreover, it has been shown that PLIN3-positive and PLIN2-positive LDs display an increase during tumor progression and especially in clear cell carcinomas [21]. Another proposed scenario suggests that most tumors are more likely to favor the use of PLIN3-positive and/or PLIN2-positive LDs [21], due to their small size [23, 24], as compared to PLIN1-posistive LDs. As a result, small LDs can be generated and consumed faster, in order to meet the energy demands of rapidly proliferating cancer cells [21].
The importance of perilipins in tumor metabolism has been poorly studied [21]. Cancer cells have the potential to exploit an aberrant lipid metabolism along with altered lipophagy. This is intensified during the extreme stress conditions of anti-cancer therapy, leading, in turn, to metabolic adaptations that would allow cancer cell survival and tumor relapse [14]. It has been suggested that lipophagy plays a significant role in turning various cell types resistant to cell death stimuli [16]. Hence, critical molecules involved in lipophagy, such as perilipins, could serve as potential targets in order to tackle the development of tumor resistance to chemotherapy or radiotherapy [25].
In the current study, we investigated the role of PLIN3 in cell viability and autophagic response of PCa cell lines treated with docetaxel. This led to the striking finding that loss of PLIN3 expression induced resistance to docetaxel. To this end, the response of the autophagic flux to docetaxel treatment was explored in PLIN3-depleted PCa cells, along with their shControl cell counterparts. Moreover, the possible involvement of apoptosis-related molecules in impaired chemotherapy treatment efficacy of PCa cells was also investigated. Finally, to investigate the importance of PLIN3 in the clinic, the expression of PLIN3 in a series of PCa tissue specimens was evaluated.
Materials and methods
Cell lines and cell culture conditions
The prostate cancer cell line 22Rv1 was purchased from the American Type Culture Collection (lgcstandards-atcc.org/Products/All/CRL-2505.aspx, Manassas, USA) and the prostate cancer cell lines DU145 and PC3 were purchased from the Cell Line Services (clsgmbh.de/p708_DU-145.html and clsgmbh.de/p1699_PC-3.html respectively, Germany). All cell lines were cultured under aseptic conditions using DMEM Low Glucose (Dulbecco's Modified Eagle Medium) (Biosera, UK), supplemented with 10% (v/v) Fetal Bovine Serum (Biosera, UK) and 1% (v/v) Penicillin/Streptomycin (Biosera, UK). Cell cultures were maintained under standard conditions at 37 °C with 5% CO2 in a humidified incubator (Heraeus, 12L01I001). Validation of the Authentication of the PCa cell lines was issued by Eurofins-Forensik (Germany).
RNA interference and development of stably transfected cell lines
All PCa cell lines were stably transfected employing a 2nd generation Lentiviral system. Two different lentiviral-based plasmid vectors were designed; one was carrying a unique sequence for the shRNA of PLIN3 and the other was free of any coding sequence. In addition, specific genes conferring resistance in two antibiotics (Puromycin & Ampicillin), a gene responsible for expressing the fluorescent mCherry protein, and a single EcoRI restriction site were also included in the aforementioned vectors. Both plasmids were purchased from GenePharma (Shanghai GenePharma, China). Each plasmid was further amplified into DH5-Alpha competent bacteria cells, followed by Ampicillin-induced selection of the successfully transformed bacterial clones. Then, plasmid DNA was extracted and purified using the NucleoSpin Plasmid Miniprep kit (REF#740,588.10, Macherey–Nagel, Germany). In order to confirm the desirable extracted plasmid product, each plasmid of interest was digested with EcoRI restriction enzyme, producing a single linear DNA molecule, detected through gel electrophoresis. Subsequently, lentiviral particles were produced after transient transfection of HEK293T host cells with the aforementioned described lentiviral vector [LV10N (U6/mCherry&Puro&Amp)], combined with the appropriate envelope plasmid (pczVSV-G) (cat#8454, Addgene) and packaging plasmid (pCMVR8.74) (cat#22,036, Addgene). HEK293T cells were transfected using Lipofectamine2000 transfection reagent (cat#11,668,019, Thermo-Fisher Scientific Inc., USA). Afterwards, the supernatant from HEK293T, containing the newly produced lentiviral particles was collected. This was followed by centrifugation at 1000 g for 10 min, filtration through a sterile filter (0.22 μm) and either storage in aliquots at − 80 °C, or immediate use to infect the PCa target cells for 48 h. Finally, the successfully transfected cells were selected in Puromycin (at a concentration of 4 μg/mL) and gradually passaged into culture. Microscopy images were acquired with the Cell IQ imaging system (× 10 magnification) in order to evaluate any morphological changes in the aforementioned PCa cell lines post stable transfection.
Treatment with docetaxel and chloroquine
Stably transfected PCa cell lines (DU145, 22Rv1 and PC3) were allowed to recover from the selection phase. All experiments were simultaneously conducted in “shPLIN3” cells (transfected with shRNA for PLIN3) and their relative “shControl” cell counterparts (transfected with non-coding sequence).
Docetaxel was purchased as an infusion solution of 20 mg/ml (Actavis Pharma, Iceland). Initially, all shControl PCa cells were exposed to escalated concentrations of docetaxel (investigated doses were 1 nM, 2 nM, 10 nM) for 24 h, in order to establish the concentration that allows approximately a mean value of more than 50% of cell viability. The optimal docetaxel dose used was 0.5 nM, 1 nM and 2 nM for DU145, 22Rv1 and PC3 cell lines, respectively.
All PCa cell lines were treated with 20 μΜ chloroquine (cat# C6628-25G, Sigma-Aldrich, USA) for 24 h. Experiments were repeated three times.
Cell viability/cytotoxicity assays
Regarding cell viability and proliferation experiments, the growth rate for each PCa cell line was determined using the resazurin-based assay AlamarBlue (cat# 88,952, Thermo-Fisher Scientific Inc., USA); as previously described [26]. Experiments were repeated three times. For all PCa cell lines, 250 cells/well were seeded in 96-wellplates in 6plicates and incubated for 24 h, in order to achieve proper cell attachment on the well surface. Subsequently, each cell line was either exposed or not to docetaxel, and cell viability was recorded 4 days later. Measurements were captured using the FLUOstar Omega microplate reader (BMG Labtech GmbH) and presented as Relative Fluorescence Units (RFUs), comprising the background fluorescent signal subtracted from the fluorescent signal of each individual well. These measurements were proportional to the living cells in each well. Correlation between the docetaxel-treated group and the untreated control group was calculated utilizing the following two equations: (a) Normalized values = RFUs at 4th day/RFUs at 1st day, and (b) Fold reduction values = Normalized values of docetaxel-treated group/Average of normalized values of the untreated group.
Western blot
For immunoblotting, all PCa cell lines were seeded in 6-well plates at a density of 3 × 105 cells/well the day prior to docetaxel treatment. Subsequently, cells were treated with different docetaxel concentrations (at 0.5 nM, 1 nM and 2 nM for DU145, 22Rv1 and PC3 respectively) 24 h before collecting the whole cell extracts. Lysates were prepared after incubation of cells in sucrose-based lysis buffer (0.25 M sucrose, 25 mM Tris–HCl, pH 7.4) with protease/phosphatase inhibitors for 30 min on ice under frequent agitation, followed by scraping and manual homogenization. Protein concentration was determined using the BCA protein assay kit (cat#23,225, Pierce, USA). Protein samples (20 μg) were resolved by discontinuous SDS-PAGE (8–12%, depending on the studied protein) and transferred onto PVDF membranes (cat# ISEQ00005, Merck-Millipore, USA) at 120 mA for 90 min at 4 °C. Subsequently, membranes were blocked in 5% non-fat dried milk for 1 h at RT, and incubated O/N at 4 °C with the appropriate rabbit primary antibodies against PLIN3 (1:1000,ab47638, Abcam, UK), LC3A (1:1000, ab62720, Abcam, UK), p62 (1:1000, ab64134, Abcam, UK), LAMP2a (1:1000, ab18528,Abcam, UK) and Caspase-9 (1:1000, ab47537,Abcam, UK). Next, membranes were incubated with anti-Rabbit HRP-conjugated secondary antibody (1:2000, BioRad, USA) for 1 h at RT, followed by detection of the chemiluminescent signal using ECL substrates (Thermo-Fisher Scientific Inc., USA). Each membrane was re-probed with mouse antibody against β-actin to confirm equally loading of samples in the SDS-PAGE (1:5000, NB 600–501, Novus Biologicals, USA). Blot images were developed using the Chemidoc® MP imaging system (BioRad, USA) and band intensity was analyzed with the ImageLab software (BioRad, USA). Experiments were repeated three times.
Immunohistochemistry
Formalin-fixed paraffin-embedded tissues of 30 surgical specimens from patients with prostate cancer, treated with radical prostatectomy were retrieved from the archives of the Department of Pathology, University Hospital of Alexandroupolis. Three μm thick tissue sections were deparaffinized, and antigen retrieval followed by heating in a dry oven for 30 min at 80 °C, in the EnVision FLEX Target Retrieval Solution pH 9.0 (DAKO, Glostrup, Denmark). Endogenous peroxidase was quenched with EnVision Flex Peroxidase Block (DAKO) for 10 min. Next, samples were incubated with the primary rabbit polyclonal anti-PLIN3 antibody (1:100, ab47638, Abcam,UK) for 1 h. Thereafter, sections were incubated with the respective secondary antibody (EnVision Flex/HRP, DAKO) for 30 min at RT. Color was developed after 5 min of incubation time with DAB solution, followed by light counterstaining with Hematoxylin QS (Cat. #H-3404, Vector Laboratories Inc., USA).
Assessment of PLIN3 expression was performed at × 200 magnification. The percentage of cells with absent, weak or strong cytoplasmic expression was recorded per each optical field and the mean value from all optical fields was used to score each case.
RNA isolation, cDNA synthesis, real time-PCR
Real time-PCR (RT-PCR) experiments were performed as previously described [27]. Briefly, total RNA samples (500 ng) were collected using the NucleoSpin RNA Plus kit (cat# 740,984.50, Macherey–Nagel, Germany), and synthesis of cDNA was performed using the PrimerScript RT Reagent Kit (cat# RR037A, TaKaRa, Japan). Expression levels of each studied gene were measured through real time qPCR, using the KAPA SYBR FAST qPCR kit (cat# KK4611, KAPA Biosystems, USA), suitable for LightCycler® 480 instrument (KR0392-v7.14, Kapa Biosystems, USA).
Specific primer sets were designed by Roche tool in order to amplify human MAP1LC3A: (FW) 5′-CATGAGCGAGTTGTTCAAGA-3′; (REV) 5′-CCATGCTGTGCTGGTTCA-3′, human SQSTM1/p62: (FW) 5′-AGCTGCCTTGTACCCACATC-3; (REV) 5′-CAGAGAAGCCCATGGACAG-3′ and human HPRT1: (FW) 5′-TGACCTTGATTTATTTTGCATACC-3′; (REV) 5′-CGAGCAAGACGTTCAGTCCT-3′. All primer sets were purchased from Roche.
The relative expression levels of autophagy related genes were determined according to the 2−ΔΔCt formula, where ΔΔCt refers to ΔCt (target)-ΔCt (reference), and ΔCt refers to the difference in threshold cycle values of either MAP1LC3A or SQSTM1/p62 to the HPRT-1 housekeeping gene. All samples were analyzed in triplicates.
ATP analysis
PCa cells were seeded 16 h prior the assay, at a density of 2 × 104 cells/well in 96-well plates compatible with luminescence detection (cat# LT27-102, Lonza, USA). The following day, cells were lysed and basal intracellular ATP levels were measured using the ATP Bioluminescence assay ViaLight™ plus kit (cat# LT07-221, Lonza, USA), according to the manufacturer’s instructions. Measurements were obtained using the FLUOstar Omega microplate reader (BMG Labtech GmbH) and presented as Relative Light Units (RLUs). Background luminescent signal was subtracted from the luminescent signal of each individual well.
Statistical analysis
Analysis was performed using the GraphPad Prism 7.0 package. Data were analyzed as continuous variables by the unpaired two-tailed- t-test with Welch’s correction. All comparisons with p value < 0.05 were considered statistically significant. Data are expressed as means ± SD. Non-significant results are not indicated in the figures.
Results
Characterization of three PCa cell lines after shRNA-mediated silencing of the PLIN3 gene
In order to assess the role of lipophagy in PCa, PLIN3 was silenced in three PCa cell lines, namely DU145, 22Rv1 and PC3. Following stable transfection with PLIN3-specific shRNA, effective silencing was confirmed (Fig. 1a). Proliferation assays indicated that all three shPLIN3 PCa cell lines exhibit a slower proliferation rate compared to scramble shRNA (shControl) (Fig. 1b). However, assessment of basal intracellular ATP levels showed that all three shPLIN3 PCa cell lines exhibited elevated ATP levels compared to their shControl counterparts (Fig. 1c). Among them, shPLIN3 PC3 cells demonstrated the highest ATP levels, followed by shPLIN3 DU145 and shPLIN3 22Rv1 cells. It is worth mentioning that upon evaluation of the microscopy images of all PCa cell lines, morphological appearance between each shPLIN3 cell line and their shControl counterparts remained unaltered (supplementary Fig. 1).
Effect of PLIN3 knockout on PCa cell viability: a Western blot analysis depicting PLIN3 stable silencing of DU145, 22Rv1 and PC3 cell lines (β-actin was used as the loading control). b Comparative graph of the proliferation rates between shControl and shPLIN3 cells for DU145, 22Rv1 and PC3 cell lines respectively, assessed with the AlamarBlue assay 4 days post seeding. Statistical significance is indicated by asterisks, *p < 0.05. c The relative basal ATP levels of sh-treated DU145, 22Rv1 and PC3 cell lines (shControl vs. shPLIN3 cells) was assessed with the ViaLight plus kit. Statistical significance is indicated by asterisks, *p < 0.05, **p < 0.01, ***p < 0.001. d Cell viability of PCa cell lines to escalated doses of docetaxel (1 nM, 2 nM and 10 nM) indicating their respective IC50 profile, 4 days post treatment, assessed with the AlamarBlue assay. The optimal docetaxel dose used was 0.5 nM, 1 nM and 2 nM for DU145, 22Rv1 and PC3 cell lines, respectively. e Response to docetaxel 4 days post treatment for shControl-DU145 vs. shPLIN3-DU145 cell lines (**p < 0.01); f shControl-22Rv1 vs. shPLIN3-22Rv1 (**p < 0.01); g shControl-PC3 vs. shPLIN3-PC3 (p = ns). Comparisons refer to shControl cells vs. shPLIN3-cells exposed to docetaxel. Bars show standard deviation and the statistically significant differences between shControl cells and shPLIN3 cells are indicated by asterisks. Results with no statistical significance are not shown in the graphs. Key labeling for the three PCa cell lines (square box) refers to Figs. 2b–g. RLUs correspond to Relative Light Units. DTX and UN correspond to docetaxel treatment and untreated cells, respectively
Differential exposure of three PCa cell lines to docetaxel treatment
All shControl PCa cell lines were exposed to escalating doses of docetaxel (1 nM, 2 nM, 10 nM). Determination of IC50 values for each PCa cell line showed that PC3 cell line was more resistant to docetaxel, compared to 22Rv1 and DU145 cell lines, with the latter one being the most sensitive (Fig. 1d).
Both shControl and shPLIN3 PCa cell lines were exposed to docetaxel. PLIN3 silencing reduced docetaxel-induced cytotoxicity in DU145 and 22Rv1 cells (p < 0.01; Fig. 1e–f respectively). shPLIN3 DU145 and shPLIN3 22Rv1 cells exhibited a onefold and 0.5-fold increase in viability compared to their shControl cells (p = 0.0057, p = 0.0002). Therefore, it seems that both shPLIN3 DU145 and shPLIN3 22Rv1 cell lines may activate or intensify survival pathways upon exposure to docetaxel that partially block docetaxel-induced cytotoxicity.
In contrast, as both shControl and shPLIN3 PC3 cells were exposed to the highest docetaxel dose and demonstrated similar sensitivity, it was suggested that PC3 cells may already possess active basal pathways that could potentially reduce docetaxel efficacy which remained unaffected upon PLIN3 gene silencing (Fig. 1g).
Comparative steady state of autophagic flux levels in untreated shPLIN3 and shControl PCa cell lines
We have previously shown that the autophagic machinery is an important mechanism involved in resistance of cancer cells to chemotherapy [28]. Therefore, we studied the involvement of autophagic pathway in the aforementioned differential response of shPLIN3 prostate cancer cell lines to docetaxel.
Initially, we examined the basal expression levels of autophagy-related proteins LC3A, LC3A-II, p62 and the lysosomal membrane protein LAMP2a in the untreated groups (shControl vs. shPLIN3 cells). Western blot analysis showed that shPLIN3 DU145 cell line exhibited higher basal levels of LC3A (0.7-fold higher), p62 (0.4-fold higher) and LAMP2a (0.5-fold higher), compared to the parental cell line, while LC3A-II levels remained unaltered (supplementary Fig. 2a). RT-PCR showed elevated levels of LC3A gene (0.5-fold higher) and a marginal increase of the p62 gene expression in shPLIN3 DU145 cell line compared with the shControl DU145 cells (supplementary Fig. 2b). The lack of LC3A-II accumulation combined with the increased gene transcription of LC3A and p62 in shPLIN3 DU145 cells suggest autophagosome consumption. Thus, shPLIN3 cells seem to have higher basal levels of autophagy flux compared to their parental cells.
A different pattern of expression was noted in the case of shPLIN3 22RV1 cell line. This cell line demonstrated higher basal protein levels of LC3A-II (0.6-fold higher) accompanied by reduced levels of p62 protein, as compared to their shControl cells (supplementary Fig. 2c). Comparison of mRNA basal levels showed no particular change in the expression of LC3A gene and a slight decrease of p62 gene (0.3-fold decrease) between shControl and shPLIN3 22Rv1 cells (supplementary Fig. 2d). Hence, no apparent effect of PLIN3 silencing on autophagic flux was noticed in untreated cells.
Regarding PC3 cell lines, PLIN3 silencing resulted in increased protein expression of LC3A-II, and LAMP2a, accompanied by reduced expression levels of p62, compared to shControl PC3 cells (supplementary Fig. 2e). Although consumption of p62 upon enhanced auto-lysosomal fusion could suggest intensified autophagic flux, RT- PCR analysis showed similar LC3A mRNA expression and decreased expression of p62 gene (0.5-fold decrease) (supplementary Fig. 2f). Thus, there was no evident alteration in the autophagic flux upon shPLIN3 silencing in PC3 cell lines.
Differential autophagic response of PCa cell lines to docetaxel
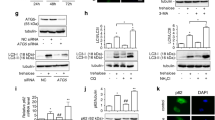
Next, we examined whether exposure to docetaxel affects the autophagic flux in PCa cells. Exposure of shControl and shPLIN3 DU145 cells to docetaxel resulted in differential autophagic responses. Soluble LC3A, membrane-bound LC3A-II and p62 proteins increased in shControl DU145 cell line (Fig. 2a). Moreover, a significant reduction of LC3A mRNA expression and a marginal reduction of the p62 mRNA levels was observed, suggesting a blockage of the autophagic flux in the shControl cell line (Fig. 2b). On the contrary, exposure of shPLIN3 DU145 cells to docetaxel was accompanied by decreased p62 and LAMP2a proteins levels, while LC3A and LC3A-II protein levels remained unchanged (Fig. 2a). Simultaneous increase of LC3A and p62 mRNA levels confirmed enhanced autophagic flux (Fig. 2b).
Expression levels of autophagy-related proteins and genes in DU145, 22Rv1 and PC3 cell lines post docetaxel treatment (shControl vs. shPLIN3 cells): a Protein expression levels of LC3A, LC3A-II, p62 and LAMP2a after exposure of shControl and shPLIN3 DU145 cells to docetaxel (0.5 nM for 24 h), b gene expression levels of LC3A and p62, after exposure of shControl and shPLIN3 DU145 cell lines to docetaxel (0.5 nM for 24 h). Statistical significance is indicated by asterisks, *p < 0.05, **p < 0.01. c Protein expression levels of LC3A, LC3A-II, p62 and LAMP2a after exposure of shControl and shPLIN3 22Rv1 cells to docetaxel (1 nM for 24 h), d gene expression levels of LC3A and p62after exposure of shControl and shPLIN3 22Rv1 cell lines to docetaxel (1 nM for 24 h). Statistical significance is indicated by asterisks, *p < 0.05. e Protein expression levels of LC3A, LC3A-II, p62 and LAMP2a after exposure of shControl and shPLIN3 PC3 cells to docetaxel (2 nM for 24 h), f gene expression levels of LC3A and p62, after exposure of shControl and shPLIN3 PC3 cell lines to docetaxel (2 nM for 24 h). Statistical significance is indicated by asterisks, *p < 0.01, **p < 0.001, ***p < 0.0001. Key labeling for the three PCa cell lines (square box) refers to the respective treatment condition of each experiment. Bars show standard deviation and results without statistical significance are excluded from the graphs. In Western blot analysis, β-actin was used as the loading control. DTX and UN correspond to docetaxel treatment and untreated cells respectively
As in shControl DU145 cell line, exposure of shControl 22Rv1 cells to docetaxel was also accompanied by increased LC3A protein levels (both soluble and membrane-bound forms), while p62 and LAMP2a protein levels remained unaltered (Fig. 2c). LC3A and p62 mRNA levels displayed a decreasing trend, suggesting a suppression of the autophagic flux upon exposure of the parental cells to docetaxel (Fig. 2d). In contrast, exposure of shPLIN3 22Rv1 cells to docetaxel, lead to a reduction of the membrane-bound LC3A and p62 protein levels (Fig. 2c), accompanied by increased LC3A and p62 mRNA levels, thus suggesting an enhancement of autophagic flux (Fig. 2d).
Treatment of shControl PC3 cells with docetaxel had no effect on LC3A, LC3A-II, p62 and LAMP2a protein levels (Fig. 2e). Regarding mRNA expression patterns, the shControl PC3 cells exhibited reduced LC3A mRNA levels and unchanged p62 levels, thus suggesting that docetaxel had no apparent effect on their autophagic flux (Fig. 2f). In contrast, exposure of shPLIN3 PC3 cells to docetaxel resulted in decreased soluble and membrane-bound LC3A, p62 and LAMP2a protein levels (Fig. 2e). Moreover, the simultaneous significant increase of LC3A and p62 mRNA levels suggested enhanced autophagic flux (Fig. 2f).
Inhibition of autophagy reverses resistance to docetaxel in PCa cells
The enhanced autophagic flux recorded in shPLIN3 PCa cell lines upon docetaxel treatment, suggested an autophagy-induced resistance. Hence, we further examined whether autophagy blockage could restore sensitivity of shPLIN3 PCa cells to docetaxel. Next, all shControl and shPLIN3 PCa cell lines were exposed to docetaxel, chloroquine (a potent autophagy flux inhibitor), and a combination thereof.
Addition of chloroquine to docetaxel had no effect on the viability of shControl DU145 cells. Interestingly, addition of chloroquine sensitized shPLIN3 cells to docetaxel, reducing their viability by 30% (Fig. 3a). Similarly, both shControl 22Rv1 and shControl PC3 cells showed unaltered viability upon concurrent exposure to docetaxel and chloroquine (Fig. 3b–c, respectively). In contrast, a strong sensitization was observed upon simultaneous treatment of both shPLIN3 22Rv1 (60% decrease) and shPLIN3 PC3 (50% decrease) cells with docetaxel and chloroquine (Fig. 3b–c, respectively). It should be noted that chloroquine per se, at the concentration applied, did not affect the viability of either DU145 or PC3 cells, while 22Rv1 cells showed a minor unexpected reduced cell viability (10% for shControl and 20% for shPLIN3 cells) upon chloroquine treatment, without any noteworthy impact to the overall result (Fig. 3a–c, respectively).
Sensitization effect of chloroquine on docetaxel cytotoxicity in PCa cell lines (shControl vs. shPLIN3 cells): a Response of shControl and shPLIN3 DU145 cells to docetaxel (0.5 nM for 24 h), chloroquine (20 μΜ for 24 h) and their combination, b response of shControl and shPLIN3 22Rv1 cells to docetaxel (1 nM for 24 h), chloroquine (20 μΜ for 24 h) and their combination, c response of shControl and shPLIN3 PC3 cells to docetaxel (2 nM for 24 h), chloroquine (20 μΜ for 24 h) and their combination. Docetaxel is designated as DTX, chloroquine as CQ, and the combined treatment as DTX+CQ. Untreated cells are shown as shControl and shPLIN3. For each treatment group all comparisons were held against their respective untreated cells
Involvement of Caspase-9 to docetaxel response
Next, we examined whether Caspase-9-driven apoptosis initiation is involved in the resistance of shPLIN3 PCa cells to docetaxel. To this end, all PCa cell lines were exposed to the aforementioned docetaxel concentrations for 24 h, 48 h, and 72 h, followed by evaluation of Caspase-9 protein levels (Fig. 4). A higher dose of docetaxel was also used for all cell lines (10 nM for 24 h). In order to assess the potential of PCa cells to initiate apoptotic response, the ratio of cleaved Caspase-9/pro-Caspase-9 was determined (Fig. 4).
Expression levels of Caspase-9 protein in all studied PCa cell lines upon treatment with docetaxel (shControl vs. shPLIN3 cells): a protein expression levels of pro-Caspase-9 and cleaved Caspase-9 after exposure of shControl and shPLIN3 DU145 cells to docetaxel (0.5 nM for 24 h, 48 h, 72 h and 10 nM for 24 h), b protein expression levels of pro-Caspase-9 and cleaved Caspase-9 after exposure of shControl and shPLIN3 22Rv1cells to docetaxel (1 nM for 24 h, 48 h, 72 h and 10 nM for 24 h), c. protein expression levels of pro-Caspase-9 and cleaved Caspase after exposure of shControl and shPLIN3 PC3 cells to docetaxel (2 nM for 24 h, 48 h, 72 h and 10 nM for 24 h). Densitometry graphs on the right side of the Fig. illustrate the apoptosis initiation potential as calculated by the ratio of cleaved Caspase-9/pro-Caspase-9 for DU145, 22Rv1, and PC3 cell lines respectively. All comparisons were held against the shControl UN for each PCa cell line, andβ-actin was used as the loading control. Treatment with docetaxel is designated as DTX, untreated cells as UN, and DTX++ refers to 10 nM docetaxel. Bars show standard deviation and statistical significance is indicated by asterisks, *p < 0.05, **p < 0.01, ***p < 0.001
The expression patterns of the apoptotic initiation were similar in shControl DU145 and shControl 22Rv1 cells in a time dependent manner. In both cell lines, apoptosis initiation, increased 72 h after exposure to docetaxel. This response was, however, significantly less intense in both shPLIN3 DU145 and shPLIN3 22Rv1 cells (Fig. 4a–b, respectively).
In PC3 cell line, the Caspase-9-driven apoptosis initiation was the least intense among the studied PCa cell lines. Moreover, the ratio of active cleaved-Caspase-9 to pro-Caspase-9 levels showed faster response at 24 h, but decreased 72 h post exposure to docetaxel in both shControl and shPLIN3 cells. Also, shPLIN3 PC3 cells displayed weaker apoptosis initiation compared to shControl cells (Fig. 4c).
Overall, shControl PCa cells demonstrated a higher potential to initiate a Caspase-9 related apoptotic response to docetaxel in a time dependent manner, compared to their shPLIN3 PCa cell counterparts.
PLIN3 expression in prostate cancer
Following the above in vitro data, suggesting PLIN3-related resistance of PCa to docetaxel chemotherapy, the question whether PLIN3 is down-regulated, at least in a subset of prostate carcinomas, was raised. Thus, we examined the expression of PLIN3 protein in a series of tissue specimens from patients treated with radical prostatectomy (Fig. 5). Immunohistochemical staining demonstrated strong expression of PLIN3 in normal prostate glands (Fig. 5a). Lack of cytoplasmic PLIN3 expression in the whole cancer cell population was recorded in 13/30 (43.4%) patients (Fig. 5b). Weak PLIN3 expression in 20–90% (median 50%) of cancer cells was noted in 7/30 patients (23.3%) (Fig. 5c), while strong PLIN3 expression was recorded in 10/30 (33.3%) patients and this concerned 10–80% (median 50%) of the total cancer cell population (Fig. 5d).
Discussion
Chemotherapy with docetaxel or cabazitaxel is the only approved therapy for metastatic castration-resistant prostate cancer [29]. Despite their approval for clinical use, their activity remains quite low. The benefit obtained by docetaxel over mitoxantrone chemotherapy is confined by a 16% increase of biochemical responses, a 2,4 month increase of median survival and a 10% increase of parameters related to the quality of life [8]. Cabazitaxel does not further improve the results obtained with docetaxel [29]. Understanding the mechanisms of resistance of prostate cancer to taxane therapy will allow the better design of clinical trials aiming to improve the efficacy of chemotherapy in this group of prostate cancer patients who cannot benefit from hormonal therapies. Mechanisms postulated to be involved in prostate cancer resistance to taxanes include overexpression of p-glycoprotein, alterations of tubulins and over-activity of various signaling pathways [30].
In the current study we unveiled an additional resistance mechanism that involves over-activation of autophagy in PLIN3-deficient cells, as a survival response of PCa cells to docetaxel. Overall, interference with the process of lipophagy via PLIN3 permanent silencing revealed four major findings: (i) reduced proliferating rate, (ii) acquired chemoresistance to docetaxel-treated cells, (iii) enhancement of the autophagic flux upon exposure to docetaxel, and (iv) reduced Caspase-9-driven apoptosis initiation. DU145 and 22Rv1 PCa cell lines depleted for PLIN3, surprisingly managed to overcome docetaxel-induced cytotoxic stress, mainly by autophagy activation, thus, resulting in reduced drug efficacy. Regarding PC3 cell lines, although both shControl and shPLIN3 cells showed a similar sensitivity to a high given dose of docetaxel (2 nM), shPLIN3 cells also demonstrated enhancement of the autophagic flux post treatment with docetaxel, similarly to the DU145 and 22Rv1 shPLIN3 cells.
Consumption of autophagy-related proteins (LC3A-II, p62, LAMP2a) and increase of mRNA levels (LC3A, p62) suggested enhanced autophagy. This is possible because during autophago-lysosome degradative/recycling machinery, autophagosomes are fused with lysosomes, where the involved proteins LC3A-II, p62 and LAMP2a are degraded by lysosomal hydrolases. LC3A and LC3A-II kinetics, are considered markers of autophagic flux [31,32,33], particularly when evaluated together with p62, a protein responsible for autophagosome formation and for carrying the ubiquitinated proteins marked for degradation. Hence, levels of LC3A-II, p62 and LAMP2a are expected to decline due to their hydrolysis upon active autophagic flux [31, 32]. In addition, increased mRNA levels of LC3A and p62 in all shPLIN3 PCa cell lines upon docetaxel treatment provide useful insights of unimpaired autophagy induction. The term ‘Autophagic flux’ refers to the effectiveness of the autophagic machinery in the cell, and is defined by the interaction of two rates: creation and consumption of autophagosomes. Consumption is the result of autophago-lysosomal fusion and digestion of the content with subsequent degradation of autophagosomes. Enhanced and sustained autophagy flux demands robust transcription rates of specific genes involved in autophagy and lysosomal genesis (i.e., LC3A, p62, LAMP2a) combined with lack of obstacles in auto-lysosomal fusion and degradation. Reduced flux can be either a result of low transcription rates of autophagy and lysosomal genesis genes, or a blockage of auto-lysosomal fusion and degradation (the latter features are often associated with high p62 protein accumulation, since p62/sequestosome protein that carries the digestible material to the autophagosomes is expected to degrade during the digestion process). Thus, accumulation of autophagosomal protein is not a reliable marker of autophagy function, as it can occur either under a robust autophagic process or blocked autophagy. Although the process of autophagy flux is complex and demands a variety of molecular and in vivo microscopy techniques to, at least partly, unveil the real autophagy state [27, 31] combined assessment of transcription and expression levels of autophagosome (e.g., LC3), lysosome (eg. LAMP2a) and p62 proteins can provide a rough index of autophagy flux status.
The dual role of autophagy, either by being pro-death or pro-survival in response to chemotherapeutic agents is widely known in the literature [34, 35]. Moreover, there is a constantly growing clinical evidence suggesting that autophagy inhibition can be achieved by targeting any of the five distinct autophagy-related stages (initiation, nucleation, elongation, maturation, degradation); especially the latter two, during which autophagosomes are fused with lysosomes, leading to the degradation of unwanted cellular components [34, 35]. Therefore, in order to determine whether the involvement of autophagy was indeed highly responsible for docetaxel reduced efficiency, all PCa cell lines were exposed to the combination of docetaxel and chloroquine, a potent inhibitor of autophagy that impairs the fusion between autophagosomes and lysosomes. Our findings indicate that upon chloroquine-induced autophagy inhibition, all PLIN3-depleted PCa cells were sensitized to docetaxel treatment, confirming the autophagy-driven chemoresistance to docetaxel.
Our findings are in agreement with other groups reporting that autophagy enhancement is indeed associated with docetaxel resistance in PCa [36, 37]. Additionally, combination of chloroquine with docetaxel, suppressed the proliferation rate of chemoresistant PCa cells [36]. Of interest, data from the clinical phase-II trial “PANDORA” indicated beneficial response of mCRPCa to docetaxel when combined with high dose of pantoprazole, a proton pump inhibitor (PPI), via subsequent inhibition of autophagy induction [38]. Another research group used a potent polypeptide-mediated complex, suitable for co-delivery of docetaxel and siRNA-based inhibitor of ATG7, a critical autophagy intermediate, for breast cancer treatment [39]. Their findings demonstrated autophagy suppression, increased apoptosis rate and improved breast cancer chemotherapy efficacy compared to docetaxel alone [39]. Moreover, they observed involvement of autophagy in docetaxel resistance, as well as diverse autophagy dependence among their studied breast cancer cell lines [39]. In a similar manner, another group managed to suppress breast cancer cells survival using docetaxel-loaded nanoparticles accompanied with chloroquine-mediated autophagy inhibition [40]. Furthermore, involvement of chloroquine and AKT inhibition to docetaxel-based treatment of cutaneous squamous cell carcinoma (cSCC) resulted in a better overall treatment response [41]. Therefore, the need for additional research and the importance of treatment modalities that combine docetaxel with an autophagy inhibitor should be underlined, in order to enhance chemotherapy effectiveness.
Since autophagy and apoptosis are tightly connected during a cytotoxic stress [34, 42], we observed reduced potential of shPLIN3 PCa cell lines to adequately initiate Caspase-9-driven apoptotic response, accompanied by enhanced autophagy. A possible scenario could involve that silencing of PLIN3, an essential lipophagy-related molecule, may lead to unexpected and adverse response upon docetaxel treatment, such as autophagy activation and reduction of apoptotic potential, protecting cancer cells from cytotoxic stimuli and promoting various events connected with tumor survival and progression. In order to overcome this adaptive event of cancer cells and ultimately their ability to resist apoptosis, we could use new synthetic or natural compounds along with chemotherapeutic drugs, or further evaluate molecules that activate or inhibit Caspase-9 expression [43]. Alternatively, in cancer cells exhibiting resistance to apoptosis, induction of autophagic cell death (ACD) using compounds that activate autophagy (e.g., rapamycin) seems to be preferable compared to autophagy inhibition [35].
Furthermore, the elevated basal ATP levels observed in all shPLIN3 PCa cells when compared to their shControl cell counterparts, could be another additional adaptive feature of reduced response to chemotherapy [44]. This metabolic adaptation of cancer cells could be reversed or blocked by utilizing inhibitors that target various deregulated glycolytic pathways, combined with inhibitors of oxidative phosphorylation [45]. This could be of high clinical impact as shutting down the aberrant ATP supply of cancer cells; chemotherapy efficacy may be significantly improved.
All our findings may have important clinical relevance as well. PLIN3 staining in tissue samples derived from patients treated with radical prostatectomy showed a striking lack of PLIN3 expression in 43.4% of the total prostate cancer tissue specimens. This loss of PLIN3 expression is in contrast to the strong expression of PLIN3 in normal prostate glands, implying a common genetic event involving PLIN3 loss and impaired lipophagy during prostate carcinogenesis. Even in patients with strong expression of PLIN3, the majority of tumors exhibited loss of PLIN3 expression in a substantial percentage of cancer cells. This particular finding, in parallel with our aforementioned in vitro results, may have an important clinical value, since a high fraction of prostate cancer patients could be effectively treated with combination of docetaxel and chloroquine. Hence, to summarize, we propose a treatment plan, where docetaxel efficiency could be improved in PCa tissues lacking or under-expressing PLIN3, with the addition of chloroquine, a widely available drug for the treatment of malaria or COVID-19 infection. Furthermore, dependent on the immunohistochemical characteristics of each tumor, combination of docetaxel treatment with autophagy inhibition can be tailored to each individual patient, which would be of great clinical importance for PCa treatment.
Conclusion
In conclusion, shPLIN3 PCa cells, characterized by impaired function of lipophagy, were able to enhance and sustain autophagic flux in response to docetaxel treatment, which seems to play a key role in chemoresistance. In this study, we demonstrated that, due to lipophagy interference, unexpected cellular adaptive events were reversed via inhibition of this autophagy-driven reparative cellular process with chloroquine. As a result, shPLIN3 PCa cells were eventually sensitized to docetaxel. To this end, our findings may pave the way to anticancer therapies that aim to combine chemotherapy with autophagy inhibition (Fig. 6). It is crucial to cautiously re-evaluate the role of the unique cancer cell metabolism and take into consideration possible cancer cell adaptations, which may account for chemoresistance, in order to improve treatment efficacy and clinical outcome of anticancer therapy.
Schematic representation of possible adaptive cellular pathways involved in PCa response to docetaxel: The chemo-naive PLIN3-deficient PCa cells (“lipophagy interference”) are characterized by reduced proliferation rate and increased basal ATP levels. Exposure to docetaxel enhances their autophagy flux response and triggers autophagy-driven chemoresistance accompanied with reduced capability of apoptosis initiation. The induced chemoresistance can be successfully reversed by combining docetaxel with chloroquine resulting in tumor regression. On the contrary, the PCa cells that normally express PLIN3 (“lipophagy unaltered”) are characterized by a higher proliferation rate and lower basal ATP levels. Treatment with docetaxel reduces autophagy flux response, increases susceptibility and enhances apoptosis initiation. These observations could also be translated in a clinical context, where at a subset of PCa tissue lacking or under-expressing PLIN3, the overall docetaxel efficacy could be augmented by adding chloroquine. Ultimately, PLIN3 immunohistochemical characterization could be used as a biomarker to treat patients by combination of docetaxel with simultaneous autophagy inhibition
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492.
Fang D, Zhou L. Androgen deprivation therapy in nonmetastatic prostate cancer patients: indications, treatment effects, and new predictive biomarkers. Asia Pac J Clin Oncol. 2019;15(3):108–20. https://doi.org/10.1111/ajco.13108.
Denham JW, Steigler A, Lamb DS, Joseph D, Turner S, Matthews J, et al. Short-term neoadjuvant androgen deprivation and radiotherapy for locally advanced prostate cancer: 10-year data from the TROG 96.01 randomised trial. Lancet Oncol. 2011;12(5):451–9. https://doi.org/10.1016/S1470-2045(11)70063-8.
Hotte SJ, Saad F. Current management of castrate-resistant prostate cancer. Curr Oncol. 2010;17(Suppl 2):S72–9. https://doi.org/10.3747/co.v17i0.718.
Kyriakopoulos CE, Chen YH, Carducci MA, Liu G, Jarrard DF, Hahn NM, et al. Chemohormonal therapy in metastatic hormone-sensitive prostate cancer: long-term survival analysis of the randomized phase III E3805 CHAARTED Trial. J Clin Oncol. 2018;36(11):1080–7. https://doi.org/10.1200/JCO.2017.75.3657.
James ND, Sydes MR, Clarke NW, Mason MD, Dearnaley DP, Spears MR, et al. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet. 2016;387(10024):1163–77. https://doi.org/10.1016/S0140-6736(15)01037-5.
Petrylak DP, Tangen CM, Hussain MH, Lara PN Jr, Jones JA, Taplin ME, et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351(15):1513–20. https://doi.org/10.1056/NEJMoa041318.
Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351(15):1502–12. https://doi.org/10.1056/NEJMoa040720.
Pienta KJ. Preclinical mechanisms of action of docetaxel and docetaxel combinations in prostate cancer. Semin Oncol. 2001;28(4 Suppl 15):3–7. https://doi.org/10.1016/s0093-7754(01)90148-4.
Mansoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B. The different mechanisms of cancer drug resistance: a brief review. Adv Pharm Bull. 2017;7(3):339–48. https://doi.org/10.15171/apb.2017.041.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. https://doi.org/10.1016/j.cell.2011.02.013.
Kounakis K, Chaniotakis M, Markaki M, Tavernarakis N. Emerging roles of lipophagy in health and disease. Front Cell Dev Biol. 2019;7:185. https://doi.org/10.3389/fcell.2019.00185.
Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. https://doi.org/10.1038/oncsis.2015.49.
Cruz ALS, Barreto EA, Fazolini NPB, Viola JPB, Bozza PT. Lipid droplets: platforms with multiple functions in cancer hallmarks. Cell Death Dis. 2020;11(2):105. https://doi.org/10.1038/s41419-020-2297-3.
Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–5. https://doi.org/10.1038/nature07976.
Liu K, Czaja MJ. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013;20(1):3–11. https://doi.org/10.1038/cdd.2012.63.
Wang CW. Lipid droplets, lipophagy, and beyond. Biochim Biophys Acta. 2016;1861(8 Pt B):793–805. https://doi.org/10.1016/j.bbalip.2015.12.010.
Kimmel AR, Sztalryd C. The perilipins: major cytosolic lipid droplet-associated proteins and their roles in cellular lipid storage, mobilization, and systemic homeostasis. Annu Rev Nutr. 2016;36:471–509. https://doi.org/10.1146/annurev-nutr-071813-105410.
Thiele C, Spandl J. Cell biology of lipid droplets. Curr Opin Cell Biol. 2008;20(4):378–85. https://doi.org/10.1016/j.ceb.2008.05.009.
Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7(5):373–8. https://doi.org/10.1038/nrm1912.
Straub BK, Herpel E, Singer S, Zimbelmann R, Breuhahn K, Macher-Goeppinger S, et al. Lipid droplet-associated PAT-proteins show frequent and differential expression in neoplastic steatogenesis. Mod Pathol. 2010;23(3):480–92. https://doi.org/10.1038/modpathol.2009.191.
Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res. 2010;51(3):468–71. https://doi.org/10.1194/jlr.R000034.
Zhang Q, Zhang P, Li B, Dang H, Jiang J, Meng L, et al. The expression of perilipin family proteins can be used as diagnostic markers of liposarcoma and to differentiate subtypes. J Cancer. 2020;11(14):4081–90. https://doi.org/10.7150/jca.41736.
Wolins NE, Brasaemle DL, Bickel PE. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006;580(23):5484–91. https://doi.org/10.1016/j.febslet.2006.08.040.
Wu X, Daniels G, Lee P, Monaco ME. Lipid metabolism in prostate cancer. Am J Clin Exp Urol. 2014;2(2):111–20.
Zachari MA, Chondrou PS, Pouliliou SE, Mitrakas AG, Abatzoglou I, Zois CE, et al. Evaluation of the alamarblue assay for adherent cell irradiation experiments. Dose Response. 2014;12(2):246–58. https://doi.org/10.2203/dose-response.13-024.Koukourakis.
Mitrakas AG, Kalamida D, Giatromanolaki A, Pouliliou S, Tsolou A, Kyranas R, et al. Autophagic flux response and glioblastoma sensitivity to radiation. Cancer Biol Med. 2018;15(3):260–74. https://doi.org/10.20892/j.issn.2095-3941.2017.0173.
Karagounis IV, Kalamida D, Mitrakas A, Pouliliou S, Liousia MV, Giatromanolaki A, et al. Repression of the autophagic response sensitises lung cancer cells to radiation and chemotherapy. Br J Cancer. 2016;115(3):312–21. https://doi.org/10.1038/bjc.2016.202.
Oudard S, Fizazi K, Sengelov L, Daugaard G, Saad F, Hansen S, et al. Cabazitaxel versus docetaxel as first-line therapy for patients with metastatic castration-resistant prostate cancer: a randomized phase III Trial-FIRSTANA. J Clin Oncol. 2017;35(28):3189–97. https://doi.org/10.1200/JCO.2016.72.1068.
Galletti G, Leach BI, Lam L, Tagawa ST. Mechanisms of resistance to systemic therapy in metastatic castration-resistant prostate cancer. Cancer Treat Rev. 2017;57:16–27. https://doi.org/10.1016/j.ctrv.2017.04.008.
Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222. https://doi.org/10.1080/15548627.2015.1100356.
Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. https://doi.org/10.4161/auto.19496.
Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–63. https://doi.org/10.1016/j.cell.2007.10.035.
Mulcahy Levy JM, Thorburn A. Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 2020;27(3):843–57. https://doi.org/10.1038/s41418-019-0474-7.
Perez-Hernandez M, Arias A, Martinez-Garcia D, Perez-Tomas R, Quesada R, Soto-Cerrato V. Targeting autophagy for cancer treatment and tumor chemosensitization. Cancers. 2019;11(10):1599. https://doi.org/10.3390/cancers11101599.
Lin JZ, Wang WW, Hu TT, Zhu GY, Li LN, Zhang CY, et al. FOXM1 contributes to docetaxel resistance in castration-resistant prostate cancer by inducing AMPK/mTOR-mediated autophagy. Cancer Lett. 2020;469:481–9. https://doi.org/10.1016/j.canlet.2019.11.014.
Hu F, Zhao Y, Yu Y, Fang JM, Cui R, Liu ZQ, et al. Docetaxel-mediated autophagy promotes chemoresistance in castration-resistant prostate cancer cells by inhibiting STAT3. Cancer Lett. 2018;416:24–30. https://doi.org/10.1016/j.canlet.2017.12.013.
Hansen AR, Tannock IF, Templeton A, Chen E, Evans A, Knox J, et al. Pantoprazole Affecting Docetaxel Resistance Pathways via Autophagy (PANDORA): Phase II Trial of High Dose Pantoprazole (Autophagy Inhibitor) with Docetaxel in Metastatic Castration-Resistant Prostate Cancer (mCRPC). Oncologist. 2019;24(9):1188–94. https://doi.org/10.1634/theoncologist.2018-0621.
Gong C, Hu C, Gu F, Xia Q, Yao C, Zhang L, et al. Co-delivery of autophagy inhibitor ATG7 siRNA and docetaxel for breast cancer treatment. J Control Release. 2017;266:272–86. https://doi.org/10.1016/j.jconrel.2017.09.042.
Zhang X, Yang Y, Liang X, Zeng X, Liu Z, Tao W, et al. Enhancing therapeutic effects of docetaxel-loaded dendritic copolymer nanoparticles by co-treatment with autophagy inhibitor on breast cancer. Theranostics. 2014;4(11):1085–95. https://doi.org/10.7150/thno.9933.
Wright TJ, McKee C, Birch-Machin MA, Ellis R, Armstrong JL, Lovat PE. Increasing the therapeutic efficacy of docetaxel for cutaneous squamous cell carcinoma through the combined inhibition of phosphatidylinositol 3-kinase/AKT signalling and autophagy. Clin Exp Dermatol. 2013;38(4):421–3. https://doi.org/10.1111/ced.12138.
Rikiishi H. Novel Insights into the Interplay between apoptosis and autophagy. Int J Cell Biol. 2012;2012:317645. https://doi.org/10.1155/2012/317645.
Kim B, Srivastava SK, Kim SH. Caspase-9 as a therapeutic target for treating cancer. Expert Opin Ther Targets. 2015;19(1):113–27. https://doi.org/10.1517/14728222.2014.961425.
Katayama M, Kawaguchi T, Berger MS, Pieper RO. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ. 2007;14(3):548–58. https://doi.org/10.1038/sj.cdd.4402030.
Kim SY. Cancer energy metabolism: shutting power off cancer factory. Biomol Ther. 2018;26(1):39–44. https://doi.org/10.4062/biomolther.2017.184.
Acknowledgements
This research work was supported by the Hellenic Foundation for Research and Innovation (H.F.R.I.) under the HFRI PhD Fellowship Grant (Fellowship Number: 904) to Ioannis Lamprou.
Author information
Authors and Affiliations
Contributions
All authors read and approved the final manuscript. IL: Data curation; Formal analysis; Funding acquisition; Validation, Investigation, Resources, Visualization, Writing-original draft, Writing-review & editing. AT: Data curation, Formal analysis, Investigation, Writing-review & editing. CK: Data curation, Formal analysis, Investigation, Writing-original draft. AGM: Data curation, Investigation. ETX: Data curation, Investigation. KK: Data curation, Investigation. IK: Methodology, Resources. CEZ: Resources, Writing-review & editing. AG: Formal analysis, Methodology, Resources, Supervision, Writing-review & editing. MIK: Conceptualization, Formal analysis, Methodology, Project administration, Resources, Supervision, Visualization, Writing-review & editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
The study was in accordance with the ethical standards of our institutional research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12032_2021_1566_MOESM1_ESM.tif
Supplementary file1 Representative images of all stably transfected PCa cell lines (shControl vs. shPLIN3 cells): Microscopy images acquired with the Cell IQ imaging system (x10 magnification), depicting the unaltered morphological appearance of DU145, 22Rv1 and PC3 PCa cell lines, respectively (TIF 16966 kb)
12032_2021_1566_MOESM2_ESM.tif
Supplementary file2 Expression levels of autophagy-related proteins and genes in three PCa cell lines (shControl vs. shPLIN3 cells): Graphs on the left column illustrate the fold change of LC3A, LC3A-II, p62 and LAMP2a protein expression for a. DU145, c. 22Rv1 and e. PC3 cell lines, respectively. Graphs on the right column illustrate the mRNA expression patterns of LC3A and p62 for b. DU145, d. 22Rv1 and f. PC3 cell lines, respectively. Untreated cells are designated as UN. Bars show standard deviation and statistical significance is indicated by asterisks *p<0.05, and **p<0.01. Key labeling for the three PCa cell lines (square box) refers to their respective untreated conditions of each experiment. Results with no statistical significance are not depicted in the graphs. Graphs of this Fig. correspond to the average of three experiments and a representative image is shown in Fig. 2 (TIF 24057 kb)
Rights and permissions
About this article
Cite this article
Lamprou, I., Tsolou, A., Kakouratos, C. et al. Suppressed PLIN3 frequently occurs in prostate cancer, promoting docetaxel resistance via intensified autophagy, an event reversed by chloroquine. Med Oncol 38, 116 (2021). https://doi.org/10.1007/s12032-021-01566-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-021-01566-y