Abstract
The pandemic caused by severe acute respiratory syndrome coronavirus 2 and the related disorder i.e. “coronavirus disease 2019” (COVID-19) has encouraged researchers to unravel the molecular mechanism of disease severity. Several lines of evidence support the impact of “cytokine storm” in the pathogenesis of severe forms of the disorder. We aimed to assess expression levels of nine cytokine coding genes in COVID-19 patients admitted in a hospital. We collected clinical data of patients from their medical reports. Then, we assessed expression of genes using real-time PCR. Expression levels of IFN-G, IL-2, IL-4, IL-6, IL-17, TGF-B, IL-8, and IL-1B were significantly higher in COVID-19 patients compared with healthy controls and in both female and male patients compared with sex-matched controls. However, expression level of TNF-A was not different between COVID-19 patients and healthy controls. Expression of none of these cytokines was different between ICU-admitted patients and other patients except for IL-6 whose expression was lower in the former group compared with the latter (ratio of means = 0.33, P value = 4.82E-02). Then, we assessed diagnostic power of cytokine coding genes in differentiating between COVID-19 patients and controls. The area under curve (AUC) values ranged from 0.94 for IFN-G to 1.0 for IL-2 and IL-1B. After combining the transcript levels of all cytokines, AUC, sensitivity, and specificity values reached 100%, 100%, and 99%, respectively. For differentiation between ICU-admitted patients and other patients, IL-4 with AUC value of 0.68 had the best diagnostic power among cytokine coding genes. Expression of none of cytokine coding genes was correlated with the available clinical/demographic data including age, gender, ICU admission, or erythrocyte sedimentation rate (ESR)/C-reactive protein (CRP) levels. This study provides further evidence for contribution of “cytokine storm” in the pathobiology of moderate/severe forms of COVID-19.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
“Severe acute respiratory syndrome coronavirus 2” (SARS-CoV-2) has been shown to cause “coronavirus disease 2019” (COVID-19) (Ghafouri-Fard et al. 2020a, b). This disorder has led to catastrophic events all over the world. Several lines of evidence support the role of “cytokine storm” in the pathogenesis of severe forms of the disorder (Tang et al. 2020; Ghafouri-Fard et al. 2020a). This term is described as induction of a cascade of auto-augmenting cytokine secretion resulting from uncontrolled host immune reaction to certain immune stimulators (Tang et al. 2020). A number of recent investigations have demonstrated elevated levels of pro-inflammatory cytokines in the critically ill COVID-19 patients compared with moderately ill ones (Tang et al. 2020; Qin et al. 2020). Induction of immune responses during the course of COVID-19 has been shown to result in the over-production of enormous quantities of inflammatory proteins, eventually leading to organ damage (Yarmohammadi et al. 2020). Upregulation of pro-inflammatory cytokines such as IL-6, IL-1β, IL-18, and TNF-α in addition to their downstream molecules has been reported in these patients (Qin et al. 2020; Perricone et al. 2020). Moreover, dysregulation of a number of other inflammatory molecules such as IL-2, IL-7, IL-10, and IFN-γ has been associated with COVID-19 (Ky and Mann 2020). Totally, many studies have shown the important impact of IL-6, IFN-γ, and TNF-α in the pathobiology of this condition and related organ damage (Yarmohammadi et al. 2020). Thus, identification of these inflammatory reactions has significance in the treatment of patients with COVID-19 infection (Rana 2020). Based on the diversity of the regulatory mechanisms of immune reactions in different ethnic groups (Quach et al. 2016), evaluation of cytokine production in the context of COVID-19 infection in each population would help in the design of appropriated therapeutic options. Therefore, in the present study, we analyzed expression levels of some cytokines including those participating in adaptive immune responses (IL-2 and IL-4) and pro-inflammatory cytokines (IFN-G, IL-1, IL-6, IL-17, and TNF-α) in a population of Iranian patients with COVID-19 in comparison with sex-matched healthy controls to unravel the importance of cytokine dysregulation in the pathogenesis of this disorder.
Methods
Study Design and Participants
This is a sex-matched case–control study involving 91 COVID-19 patients (cases) and 96 healthy controls. Female/male ratios were 38/53 and 39/57 in cases and controls, respectively. The present investigation was performed on patients admitted to Nikan Hospital, Tehran, during March 2020 until April 2020. Patients with clinical symptoms of COVID-19 were further assessed through RT-PCR assay on the obtained nasopharyngeal samples. Only those with confirmed molecular diagnosis were enrolled in the study. This patient cohort included patients with moderate, severe, and very severe disease courses (Son et al. 2021). Control samples were obtained from healthy individuals without no clinical symptom or exposure to the affected individuals. The study protocol was approved by ethics committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.RETECH.REC.1399.046). Informed consent was obtained from all patients. A complete paraclinical investigation and chest CT scans were done for all patients. Hematological tests and C-reactive protein (CRP) levels were assessed using Beckman Coulter MAXM AL Hematology Flow Cytometry System and latex-enhanced nephelometry systems, respectively.
Expression Assays
As the initial step, 3–5 mL of peripheral blood was gathered from all patients and controls. Then, total RNA was recovered from these specimens using the GeneAll RNA extraction Kit (Seoul, South Korea). Subsequently, total RNA was converted to cDNA using the OneStep RT-PCR Series Kit (BioFact™, Seoul, South Korea). Expression levels of cytokine coding genes were quantified in samples obtained from COVID-19 patients and controls using the RealQ Plus 2 × Master Mix (Amplicon, Denmark) using the transcript levels of HPRT1 gene as normalizer. Reactions were performed in duplicate. PCR program consisted of 10 min initial denaturation at 95 °C, 32 cycles of denaturation at 95 °C for 5 s and annealing at 60 °C for 20 s. Table 1 displays the features of primers, probes, and amplified segments.
Statistical Methods
Statistical comparisons were performed using R programming language. Transcript quantities of nine cytokine coding genes were measured from threshold cycle (Ct) and PCR efficiency (amp) values, considering HPRT1 as the reference gene. The following formula was used: \(\frac{am{p}_{g}^{-C{T}_{g}}}{am{p}_{HPRT1}^{-C{T}_{HPRT1}}}\) where g indicates any of the cytokine coding genes. Then, the calculated values were log2 transformed and used for succeeding analyses. Two comparisons were done and the significance of difference in mean values between two subgroups was computed using the t-test. Expression levels of genes were compared between total patients and healthy controls as well as between patients who required admission in intensive care unit (ICU) and who did not. Receiver operating characteristic (ROC) curves were plotted using Bayesian generalized linear model, generalized linear model, and linear discriminant analysis with tenfold cross validation. The linear discriminant analysis (LDA) provided the most efficient estimates and in the best setting, the area under curve (AUC) value was 1. Youden’s J statistic was employed to find the optimum threshold. LDA was then chosen according to the former steps to investigate efficiency of each gene for separating groups. Spearman correlation coefficients were employed to assess the association between patients’ information and transcript levels. To compute correlation between categorical and continuous variables, point biserial correlation coefficient was used. For all statistical tests, the level of significance was set at P value < 0.05.
Results
The mean age (standard deviation) of the patients was 57.17 (16.90) years. Controls were matched with cases in their age (mean ± standard deviation = 56.25 ± 15.86). Among the admitted patients, 37 patients (40.6%) were admitted in the ICU. Table 2 shows the paraclinical parameters of the COVID-19 group.
Expression Assays
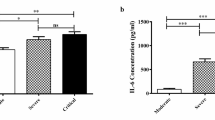
Expression levels of IFN-G, IL-2, IL-4, IL-6, IL-17, TGF-B, IL-8, and IL-1B were significantly higher in COVID-19 patients compared with controls and in both female and male patients compared with sex-matched controls. However, expression of none of these cytokines was different between ICU-admitted patients and other patients except for IL-6 whose expression was lower in the former group compared with the latter (ratio of means = 0.33, P value = 4.82E-02). Expression of TNF-A was not different between COVID-19 patients and healthy subjects (Table 3).
Figure 1 A graphical representation of relative amounts of cytokine coding genes in COVID-19 patients and controls. Minimum, maximum, median, first and third quartiles are shown.
Expression of none of cytokine coding genes was correlated with the assessed clinical/demographic data including age, gender, ICU admission, or CRP/ESR levels (Fig. 2).
Then, we assessed diagnostic power of cytokine coding genes in diagnosing between COVID-19 patients and healthy controls. AUC values ranged from 0.94 for IFN-G to 1.0 for IL-2 and IL-1B. After combining the transcript levels of all cytokines, AUC, sensitivity, and specificity values reached 1.0, 1.0, and 0.99, respectively. For differentiation between ICU-admitted patients and other patients, IL-4 with AUC value of 0.68 had the best diagnostic power among cytokine coding genes (Table 4). Figures 3 and 4 show the ROC curves depicted by three machine learning models and the best fitted one, respectively.
Lastly, we measured the correlation between the transcript levels of cytokine coding genes among ICU-admitted COVID-19 patients, non-ICU-admitted COVID-19 patients and healthy controls (Fig. 5). In the first group, the most significant correlation was found between TNF-A and TGF-B (r = 0.91, P value = 9.37E-15). In non-ICU admitted patients and healthy controls, the most significant correlations were demonstrated between TNF-A and IFN-G (r = 0.91, P value = 1.30E-21 and r = 0.92, P value = 7.90E-41, respectively).
Discussion
An essential and continuing mission in COVID-19 investigations is to find why and how SARS-CoV-2 provokes heterogeneous reactions in terms of severity and immunopathology (Buszko et al. 2021). Cytokines have important roles in the pathoetiology of COVID-19, resolution of the illness, and the effectiveness of antiviral and immunomodulatory agents (Buszko et al. 2021). Meanwhile, ethnic factors have been shown to affect susceptibility to COVID-19 and outcome of the disorder (Sze et al. 2020). Such roles of ethnic-based factors have in determination of adverse COVID-19 outcomes have been prominent even after adjustment of the sociodemographic and clinical parameters (Mathur et al. 2021). We examined expression levels of nine cytokine-coding genes among ICU-admitted COVID-19 patients, non-ICU-admitted ones, and healthy subjects. We detected over-expression of IFN-G, IL-2, IL-4, IL-6, IL-17, TGF-B, IL-8, and IL-1B in COVID-19 patients compared with healthy subjects and in both female and male patients compared with sex-matched controls. However, expression of none of these cytokines was different between ICU-admitted patients and other patients except for IL-6 whose expression was lower in the former group compared with the latter. Expression of TNF-A was not different between COVID-19 patients and healthy subjects.
Levels of several cytokines have been assessed in different subgroups of COVID-19 patients. For instance, Chen et al. have reported remarkable over-expression of IL-2R and IL-6 in the critically ill COVID-19 patients compared with severe and mild groups (Chen et al. 2020b). However, we detected a trend toward under-expression of IL-6 in the ICU-admitted patients compared with the other group of patients. This finding is not reliable as sex-based comparisons did not verify the difference in the expression of this cytokine between ICU-admitted and non-ICU admitted subjects. Moreover, this finding is in contrast with our previous study which demonstrated higher median levels of IL-6 protein in the serum of ICU-admitted patients (Samsami et al. 2020). However, it is worth mentioning that the current study varies with our previous study in the terms of applied technique and source of expression assessment. We recommend assessment of expression of IL-6 in larger cohorts of Iranian patients to unravel the possible difference in its expression between Iranian patients and patients from other populations. In accordance with our finding regarding similar levels of TNF-α, IL-1, and IL-8 cytokines between ICU-admitted and non-ICU-admitted COVID-19 patients, Chen et al. did not detect difference in the serum levels of among the critical, severe, and moderate groups COVID-19 patients (Chen et al. 2020b).
A recent study in Erbil city of Iraq has revealed no significant difference in levels of IL-1 and TNF-α between normal controls, moderately affected COVID-19 patients, severely affected COVID-19 patients, and those who recovered from this disorder. Nevertheless, authors have detected higher concentrations of IL-6 in moderately or severely affected COVID-19 cases compared to controls and recovered patients. Moreover, recovered patients had lower levels of IFN-γ and IL-4 while higher levels of IL-10 compared with severely affected cases (Merza et al. 2021).
The observed over-expression of IFN-G in COVID-19 patients in the current study is in line with the recently reported augmented nucleoprotein-induced IFN-γ release in these patients (Thijsen et al. 2020). Hu et al. have demonstrated lower probability of lung fibrosis at discharge in patients who had higher baseline levels of IFN-γ (Hu et al. 2020). Although we did not assess the presence of lung fibrosis in the admitted patients, we demonstrated similar levels of IFN-G between ICU-admitted and non-ICU-admitted patients. On the other hand, in a small cohort of Chinese patients, severely affected COVID-19 cases exhibited lower levels of IFN-γ expression by CD4 + T cells compared with moderately affected cases (Chen et al. 2020a).
Over-expression of IL-17 in COVID-19 patients has also been reported in other populations (Huang et al. 2020). In addition, MERS-CoV has been shown to stimulate expression of this cytokine in humans (Faure et al. 2014). Consistently, Th17 cells have been reported to participate in the cytokine storm stimulated by SARS-CoV-2 (Huang et al. 2020). Upregulation of IL-17, IL-2, and IL-4 levels has also been reported in COVID-19 patients with lung lesions (Huang et al. 2020).
Similar levels of TNF-A between three study subgroups in the current investigation raise the possibility of ethnic-based differences in the immunological responses in the context of COVID-19 infection, since this cytokine has been repeatedly reported to be increased in these patients and has been suggested as a target of immune-modulatory options (Costela-Ruiz et al. 2020). SARS-CoV-2 has also been suggested to activate of IL-1β, which consecutively induces other pro-inflammatory cytokines, such as IL-6 and TNF-α (Costela-Ruiz et al. 2020). However, while we detected over-expression of IL-1B and IL-6 in COVID-19 patients, we could not demonstrate any significant difference in the expression of TNF-A.
Then, we measured diagnostic power of cytokine transcripts in diagnosing COVID-19 patients from healthy controls. The AUC value was the highest for IL-2 and IL-1B. After combining the transcript levels of all cytokines, AUC, sensitivity, and specificity values reached 100%, 100%, and 99%, respectively. Therefore, cytokine levels can be used for distinguishing disease status of COVID-19 patients. In spite of suitable diagnostic power of cytokine levels for differentiation between total COVID-19 patients and healthy subjects, these molecules cannot differentiate ICU-admitted patients from non-ICU-admitted patients. Besides, expression of none of cytokine coding genes was correlated with the assessed clinical/demographic data including age, gender, ICU admission, or CRP/ESR levels. Lastly, we evaluated the correlation between the transcript levels of cytokine coding genes among three study subgroups. Patterns of correlation between expression levels of genes were more similar between non-ICU admitted patients and healthy controls, implying the altered regulatory mechanisms of cytokines expression in severely affected patients.
In summary, we demonstrated altered levels of several cytokine coding genes in Iranian patients with COVID-19 infection. This study provides further evidence for contribution of “cytokine storm” in the pathogenesis of moderate/severe forms of COVID-19. We suggest conduction of further studies to measure protein levels of these cytokines in the serum samples to confirm the results of current study.
Availability of Data and Materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
References
Buszko M, Nita-Lazar A, Park JH, Schwartzberg PL, Verthelyi D, Young H, Rosenberg AS (2021) Lessons learned: new insights on the role of cytokines in COVID-19. Nature Publishing Group
Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, Zhang X, Zhang M, Wu S, Song J, Chen T, Han M, Li S, Luo X, Zhao J, Ning Q (2020a) Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 130:2620–2629
Chen L, Liu H, Liu W, Liu J, Liu K, Shang J, Deng Y, Wei S (2020b) Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia. Zhonghua jie he he hu xi za zhi= Zhonghua jiehe he huxi zazhi= Chinese journal of tuberculosis and respiratory diseases, 43, E005-E005
Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodríguez L (2020) SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev 54:62–75
Eftekharian MM, Ghafouri-Fard S, Noroozi R, Omrani MD, Arsang-Jang S, Ganji M, Gharzi V, Noroozi H, Komaki A, Mazdeh M, Taheri M (2018) Cytokine profile in autistic patients. Cytokine 108:120–126
Faure E, Poissy J, Goffard A, Fournier C, Kipnis E, Titecat M, Bortolotti P, Martinez L, Dubucquoi S, Dessein R, Gosset P, Mathieu D, Guery B (2014) Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside? PLoS One 9:e88716
Ghafouri-Fard S, Noroozi R, Omrani MD, Branicki W, Pośpiech E, Sayad A, Pyrc K, Łabaj PP, Vafaee R, Taheri M, Sanak M (2020a) Angiotensin converting enzyme: a review on expression profile and its association with human disorders with special focus on SARS-CoV-2 infection. Vasc Pharmacol 130
Ghafouri-Fard S, Noroozi R, Vafaee R, Branicki W, Pośpiech E, Pyrc K, Łabaj PP, Omrani MD, Taheri M, Sanak M (2020b) Effects of host genetic variations on response to, susceptibility and severity of respiratory infections. Biomed Pharmacother 128
Hu Z-J, Xu J, Yin J-M, Li L, Hou W, Zhang L-L, Zhou Z, Yu Y-Z, Li H-J, Feng Y-M (2020) Lower circulating interferon-gamma is a risk factor for lung fibrosis in COVID-19 patients. Front Immunol 11:2348
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506
Ky B, Mann DL (2020) COVID-19 Clinical trials: a primer for the cardiovascular and cardio-oncology communities. JACC: Basic to Translational Science 5:501–517
Mathur R, Rentsch CT, Morton CE, Hulme WJ, Schultze A, Mackenna B, Eggo RM, Bhaskaran K, Wong AY, Williamson EJ (2021) Ethnic differences in SARS-CoV-2 infection and COVID-19-related hospitalisation, intensive care unit admission, and death in 17 million adults in England: an observational cohort study using the OpenSAFELY platform. The Lancet 397:1711–1724
Merza MY, Hwaiz RA, Hamad BK, Mohammad KA, Hama HA, Karim AY (2021) Analysis of cytokines in SARS-CoV-2 or COVID-19 patients in Erbil city, Kurdistan Region of Iraq. Plos One 16:e0250330
Perricone C, Triggianese P, Bartoloni E, Cafaro G, Bonifacio AF, Bursi R, Perricone R, Gerli R (2020) The anti-viral facet of anti-rheumatic drugs: lessons from COVID-19. J Autoimmun 102468
Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, Xie C, Ma K, Shang K, Wang W (2020) Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clinical Infectious Diseases
Quach H, Rotival M, Pothlichet J, Loh YHE, Dannemann M, Zidane N, Laval G, Patin E, Harmant C, Lopez M (2016) Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell 167:643–656. e17
Rana MM (2020) Cytokine storm in COVID-19: Potential therapeutics for immunomodulation. Journal of Research in Clinical Medicine 8:38–38
Samsami M, Mehravaran E, Tabarsi P, Javadi A, Arsang-Jang S, Komaki A, Taheri M, Ghafouri-Fard S (2020) Clinical and demographic characteristics of patients with COVID-19 infection: statistics from a single hospital in Iran. Human Antibodies 1–6
Son K-B, Lee T-J, Hwang S-S (2021) Disease severity classification and COVID-19 outcomes, Republic of Korea. Bull World Health Organ 99:62
Sze S, Pan D, Nevill CR, Gray LJ, Martin CA, Nazareth J, Minhas JS, Divall P, Khunti K, Abrams KR (2020) Ethnicity and clinical outcomes in COVID-19: a systematic review and meta-analysis. EClinicalMedicine 100630
Tang Y, Liu J, Zhang D, Xu Z, Ji J, Wen C (2020) Cytokine storm in COVID-19: the current evidence and treatment strategies. Front Immunol 11:1708–1708
Thijsen S, Heron M, Gremmels H, van der Kieft R, Reusken C, Kremer K, Limonard G, Bossink A (2020) Elevated nucleoprotein-induced interferon-γ release in COVID-19 patients detected in a SARS-CoV-2 enzyme-linked immunosorbent spot assay. J Infect 81:452–482
Yarmohammadi A, Yarmohammadi M, Fakhri S, Khan H (2020) Targeting pivotal inflammatory pathways in COVID-19: a mechanistic review. Eur J Pharmacol 173620
Author information
Authors and Affiliations
Contributions
SGF and MT wrote the manuscript and contributed in study design. MS, RJK, and HRK collected the data and confirmed the patients’ diagnosis. SHS and BMH analyzed the data. All authors read and approved the submitted manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participant
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent forms were obtained from all study participants. Informed consent forms were obtained from all study participants. The study protocol was approved by the ethical committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.MSP.REC.1399.046). All methods were performed in accordance with the relevant guidelines and regulations.
Consent of Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Samsami, M., Fatemi, A., Jalili Khoshnoud, R. et al. Abnormal Transcript Levels of Cytokines Among Iranian COVID-19 Patients. J Mol Neurosci 72, 27–36 (2022). https://doi.org/10.1007/s12031-021-01941-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-021-01941-4