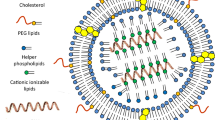
Abstract
Around 2200 miRNA (microRNA) genes were found in the human genome. miRNAs are arranged in clusters within the genome and share the same transcriptional regulatory units. It has been revealed that approximately 50% of miRNAs elucidated in the genome are transcribed from non-protein-coding genes, and the leftover miRNAs are present in the introns of coding sequences. We are now approaching a stage in which miRNA diagnostics and therapies can be established confidently, and several commercial efforts are underway to carry these innovations from the bench to the clinic. MiRNAs control many of the significant cellular activities such as production, differentiation, growth, and metabolism. Particularly in the immune system, miRNAs have emerged as a crucial biological component during diseased state and homeostasis. miRNAs have been found to regulate inflammatory responses and autoimmune disorders. Moreover, each miRNA targets multiple genes simultaneously, making miRNAs promising tools as diagnostic biomarkers and as remedial targets. Still, one of the major obstacles in miRNA-based approaches is the achievement of specific and efficient systemic delivery of miRNAs. To overcome these challenges, nanoformulations have been synthesized to protect miRNAs from degradation and enhance cellular uptake. The current review deals with the miRNA-mediated regulation of the recruitment and activation of immune cells, especially in the tumor microenvironment, viral infection, inflammation, and autoimmunity. The nano-based miRNA delivery modes are also discussed here, especially in the context of immune modulation.




Similar content being viewed by others
Data availability
Not applicable.
Code availability
Not applicable.
References
Shu J, Silva BVRE, Gao T, Xu Z, Cui J. Dynamic and modularized microRNA regulation and its implication in human cancers. Sci Rep. 2017;7:13356. https://doi.org/10.1038/s41598-017-13470-5.
Lee H-M, Nguyen DT, Lu L-F. Progress and challenge of microRNA research in immunity. Front Genet. 2014;5:178. https://doi.org/10.3389/fgene.2014.00178.
Fernández-Messina L, Gutiérrez-Vázquez C, Rivas-García E, Sánchez-Madrid F, de la Fuente H. Immunomodulatory role of microRNAs transferred by extracellular vesicles. Biol Cell. 2015;107:61–77. https://doi.org/10.1111/boc.201400081.
Desi N, Tay Y. The butterfly effect of RNA alterations on transcriptomic equilibrium. Cells. 2019;8:1634. https://doi.org/10.3390/cells8121634.
Fan J, Feng Y, Zhang R, et al. A simplified system for the effective expression and delivery of functional mature microRNAs in mammalian cells. Cancer Gene Ther. 2020;27:424–37. https://doi.org/10.1038/s41417-019-0113-y.
Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–66. https://doi.org/10.1261/rna.7135204.
Ketting RF. A dead end for microRNAs. Cell. 2007;131:1226–7. https://doi.org/10.1016/j.cell.2007.12.004.
Pradhan AK, Bhoopathi P, Talukdar S, et al. MDA-7/IL-24 regulates the miRNA processing enzyme DICER through downregulation of MITF. Proc Natl Acad Sci U S A. 2019;116:5687–92. https://doi.org/10.1073/pnas.1819869116.
Nguyen TA, Jo MH, Choi Y-G, et al. Functional anatomy of the human microprocessor. Cell. 2015;161:1374–87. https://doi.org/10.1016/j.cell.2015.05.010.
Gareev I, Beylerli O, Yang G, et al. The current state of miRNAs as biomarkers and therapeutic tools. Clin Exp Med. 2020;20:349–59. https://doi.org/10.1007/s10238-020-00627-2.
Roberts TC. The microRNA biology of the mammalian nucleus. Mol Ther–Nucleic Acids. 2014;3:e188. https://doi.org/10.1038/mtna.2014.40.
Yao Q, Chen Y, Zhou X. The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol. 2019;51:11–7. https://doi.org/10.1016/j.cbpa.2019.01.024.
Michlewski G, Cáceres JF. Post-transcriptional control of miRNA biogenesis. RNA. 2019;25:1–16. https://doi.org/10.1261/rna.068692.118.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. https://doi.org/10.1016/j.cell.2009.01.002.
Lee SWL, Paoletti C, Campisi M, et al. MicroRNA delivery through nanoparticles. J Control Release. 2019;313:80–95.
O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;3(9):402. https://doi.org/10.3389/fendo.2018.00402.
Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics. 2014;2014:970607. https://doi.org/10.1155/2014/970607.
Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–83. https://doi.org/10.1038/sj.emboj.7601512.
Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–35. https://doi.org/10.1016/j.jmb.2004.03.065.
Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. https://doi.org/10.1038/nrg2936.
Ipsaro JJ, Joshua-Tor L. From guide to target: molecular insights into eukaryotic RNA-interference machinery. Nat Struct Mol Biol. 2015;22:20–8. https://doi.org/10.1038/nsmb.293.
Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7(4):147–54. https://doi.org/10.1016/S1672-0229(08)60044-3.
Xu W, San Lucas A, Wang Z, Liu Y. Identifying microRNA targets in different gene regions. BMC Bioinformatics. 2014;15(Suppl. 7):S4. https://doi.org/10.1186/1471-2105-15-S7-S.
Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci USA. 2008;105:14879–84. https://doi.org/10.1073/pnas.0803230105.
Zhang J, ZhouW Liu Y, Liu T, Li C, Wang L. Oncogenic role of microRNA- 532–5p in human colorectal cancer via targeting of the 5′ UTR of RUNX3. Oncol Lett. 2018;15:7215–20. https://doi.org/10.3892/ol.2018.8217.
Dharap A, Pokrzywa C, Murali S, Pandi G, Vemuganti R. MicroRNA miR-324-3p induces promoter-mediated expression of RelA gene. PLoS ONE. 2013;8(11):e79467. https://doi.org/10.1371/journal.pone.0079467.
Miao L, Yao H, Li C, Pu M, Yao X, Yang H, et al. A dual inhibition: microRNA-552 suppresses both transcription and translation of cytochrome P450 2E1. Biochim Biophys Acta. 2016;1859:650–62. https://doi.org/10.1016/j.bbagrm.2016.02.016.
Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014;6:211–21. https://doi.org/10.1016/j.celrep.2013.12.013.
Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309(5740):1573–6. https://doi.org/10.1126/science.1115079.
Barman B, Bhattacharyya SN. mRNA targeting to endoplasmic reticulum precedes ago protein interaction and microRNA (miRNA)-mediated translation repression in mammalian cells. J Biol Chem. 2015;290(41):24650–6. https://doi.org/10.1074/jbc.C115.661868.
Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–9. https://doi.org/10.1038/ncb1929.
Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, Gidrol X. PremicroRNA and mature microRNA in human mitochondria. PLoS ONE. 2011;6:e20220. https://doi.org/10.1371/journal.pone.0020220.
Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–19. https://doi.org/10.1016/j.cell.2014.05.047.
Nishi K, Takahashi T, Suzawa M, Miyakawa T, Nagasawa T, Ming Y, et al. Control of the localization and function of a miRNA silencing component TNRC6A by Argonaute protein. Nucleic Acids Res. 2015;43:9856–73. https://doi.org/10.1093/nar/gkv1026.
Raisch J. Role of microRNAs in the immune system, inflammation and cancer. World J Gastroenterol. 2013;19:2985. https://doi.org/10.3748/wjg.v19.i20.2985.
O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–22. https://doi.org/10.1038/nri2708.
Janeway CA Jr, Travers P, Walport M, Capra JD. Immunobiology: the immune system in health and disease, 5th ed.New York: Garland Science; 2001.
Rossetti S, Sacchi N. RUNX1: a microRNA hub in normal and malignant hematopoiesis. Int J Mol Sci. 2013;14:1566–88. https://doi.org/10.3390/ijms14011566.
Hortu HO, Karaca E, Sozeri B, et al. Evaluation of the effects of miRNAs in familial Mediterranean fever. Clin Rheumatol. 2019;38:635–43. https://doi.org/10.1007/s10067-017-3914-0.
Meira-Strejevitch CS, Pereira I de S, Hippólito DDC, et al. Ocular toxoplasmosis associated with up-regulation of miR-155–5p/miR-29c-3p and down-regulation of miR-21–5p/miR-125b-5p. Cytokine. 2020;127:154990. https://doi.org/10.1016/j.cyto.2020.154990.
Georgantas RW, Hildreth R, Morisot S, et al. CD34 hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104:2750–5. https://doi.org/10.1073/pnas.0610983104.
Zhao JL, Rao DS, Boldin MP, et al. NF-κB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc Natl Acad Sci U S A. 2011;108:9184–9. https://doi.org/10.1073/pnas.1105398108.
Duroux-Richard I, Roubert C, Ammari M, et al. miR-125b controls monocyte adaptation to inflammation through mitochondrial metabolism and dynamics. Blood. 2016;128:3125–36. https://doi.org/10.1182/blood-2016-02-697003.
Le MTN, Shyh-Chang N, Khaw SL, et al. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genet. 2011;7:e1002242. https://doi.org/10.1371/journal.pgen.1002242.
Bazzoni F, Rossato M, Fabbri M, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A. 2009;106:5282–7. https://doi.org/10.1073/pnas.0810909106.
Li Q-J, Chau J, Ebert PJR, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–61. https://doi.org/10.1016/j.cell.2007.03.008.
Kuo G, Wu C-Y, Yang H-Y. MiR-17-92 cluster and immunity. J Formos Med Assoc. 2019;118:2–6. https://doi.org/10.1016/j.jfma.2018.04.013.
Smith NL, Wissink EM, Grimson A, Rudd BD. MiR-150 regulates differentiation and cytolytic effector function in CD8+ T cells. Sci Rep. 2015;5:16399. https://doi.org/10.1038/srep16399.
Lee J, Park H, Eom J, Kang SG. MicroRNA-mediated regulation of the development and functions of follicular helper T cells. Immune Netw. 2018;18:e7. https://doi.org/10.4110/in.2018.18.e7.
Yu D, Rao S, Tsai LM, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–68. https://doi.org/10.1016/j.immuni.2009.07.002.
Takahashi H, Kanno T, Nakayamada S, et al. TGF-β and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat Immunol. 2012;13:587–95. https://doi.org/10.1038/ni.2286.
Baumjohann D, Kageyama R, Clingan JM, et al. The microRNA cluster miR-17 92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat Immunol. 2013;14:840–8. https://doi.org/10.1038/ni.2642.
Lu L-F, Boldin MP, Chaudhry A, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–29. https://doi.org/10.1016/j.cell.2010.08.012.
Lu L-F, Thai T-H, Calado DP, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. https://doi.org/10.1016/j.immuni.2008.11.010.
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. https://doi.org/10.1038/nature03702.
Liu Y, Zhao Q, Xi T, Zheng L, Li X. MicroRNA-9 as a paradoxical but critical regulator of cancer metastasis: implications in personalized medicine. Genes Dis. 2020. https://doi.org/10.1016/j.gendis.2020.10.005.
Shin JH, Shin DH, Kim JS. Let-7 miRNA and CDK4 siRNA co-encapsulated in herceptin-conjugated liposome for breast cancer stem cells. Asian J Pharm Sci. 2020;15:472–81. https://doi.org/10.1016/j.ajps.2019.03.001.
Li Y, Zhang Y, Zou Y, Duan S. The paradoxical roles of miR-4295 in human cancer: implications in pathogenesis and personalized medicine. Genes Dis. 2020. https://doi.org/10.1016/j.gendis.2020.09.007.
Fujita Y, Kojima K, Ohhashi R, et al. MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J Biol Chem. 2010;285:19076–84. https://doi.org/10.1074/jbc.M109.079525.
Hirata Y, Murai N, Yanaihara N, et al. MicroRNA-21 is a candidate driver gene for 17q23-25 amplification in ovarian clear cell carcinoma. BMC Cancer. 2014;14:799. https://doi.org/10.1186/1471-2407-14-799.
Li H, Yuan S-M, Yang M, et al. High intensity focused ultrasound inhibits melanoma cell migration and metastasis through attenuating microRNA-21-mediated PTEN suppression. Oncotarget. 2016;7:50450–60. https://doi.org/10.18632/oncotarget.10433.
Wang F, Liu M, Li X, Tang H. MiR-214 reduces cell survival and enhances cisplatin-induced cytotoxicity via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett. 2013;587:488–95. https://doi.org/10.1016/j.febslet.2013.01.016.
Zhang H, Luo X-Q, Feng D-D, et al. Upregulation of microRNA-125b contributes to leukemogenesis and increases drug resistance in pediatric acute promyelocytic leukemia. Mol Cancer. 2011;10:108. https://doi.org/10.1186/1476-4598-10-108.
Arrieta VA, Cacho-Diaz B, Zhao J, et al. The possibility of cancer immune editing in gliomas A critical review. Oncoimmunology. 2018;7:e1445458. https://doi.org/10.1080/2162402X.2018.1445458.
Omar HA, El-Serafi AT, Hersi F, et al. Immunomodulatory microRNAs in cancer: targeting immune checkpoints and the tumor microenvironment. FEBS J. 2019;286:3540–57. https://doi.org/10.1111/febs.15000.
Kuninty PR, Schnittert J, Storm G, Prakash J. MicroRNA targeting to modulate tumor microenvironment. Front Oncol. 2016;19(6):3. https://doi.org/10.3389/fonc.2016.00003.
Wang B, Wang Q, Wang Z, Jiang J, et al. Metastatic consequences of immune escape from NK cell cytotoxicity by human breast cancer stem cells. Cancer Res. 2014;74(20):5746–57. https://doi.org/10.1158/0008-5472.CAN-13-2563.
Cheung IY, Farazi TA, Ostrovnaya I, et al. Deep MicroRNA sequencing reveals downregulation of miR-29a in neuroblastoma central nervous system metastasis. Genes Chromosomes Cancer. 2014;53(10):803–14. https://doi.org/10.1002/gcc.22189.
Gonzalez Plaza JJ. Current roles of microRNAs in infectious diseases—advancing into healthcare. Infektološki glasnik. 2016;36:5–15.
Maudet C, Mano M, Eulalio A. MicroRNAs in the interaction between host and bacterial pathogens. FEBS Lett. 2014;588:4140–7. https://doi.org/10.1016/j.febslet.2014.08.002.
Jopling CL, Yi M, Lancaster AM, et al. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–81. https://doi.org/10.1126/science.1113329.
Machlin ES, Sarnow P, Sagan SM. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci U S A. 2011;108:3193–8. https://doi.org/10.1073/pnas.1012464108.
Sarnow P, Sagan SM. Unraveling the mysterious interactions between hepatitis C virus RNA and liver-specific microRNA-122. Annu Rev Virol. 2016;3:309–32. https://doi.org/10.1146/annurev-virology-110615-042409.
Cameron JE, Yin Q, Fewell C, et al. Epstein-Barr virus latent membrane protein 1 induces cellular microRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol. 2008;82:1946–58. https://doi.org/10.1128/JVI.02136-07.
Lind EF, Elford AR, Ohashi PS. Micro-RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J Immunol. 2013;190:1210–6. https://doi.org/10.4049/jimmunol.1202700.
Gracias DT, Stelekati E, Hope JL, et al. The microRNA miR-155 controls CD8+ T cell responses by regulating interferon signaling. Nat Immunol. 2013;14:593–602. https://doi.org/10.1038/ni.2576.
Sullivan CS, Grundhoff AT, Tevethia S, et al. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–6. https://doi.org/10.1038/nature03576.
Ceribelli A, Yao B, Dominguez-Gutierrez PR, Chan EKL. Lupus T cells switched on by DNA hypomethylation via microRNA? Arthritis Rheum. 2011;63:1177–81. https://doi.org/10.1002/art.30192.
Junker A, Hohlfeld R, Meinl E. The emerging role of microRNAs in multiple sclerosis. Nat Rev Neurol. 2011;7:56–9. https://doi.org/10.1038/nrneurol.2010.179.
Li L, Zuo X, Liu D, et al. The profiles of miRNAs and lncRNAs in peripheral blood neutrophils exosomes of diffuse cutaneous systemic sclerosis. J Dermatol Sci. 2020;98:88–97. https://doi.org/10.1016/j.jdermsci.2020.02.009.
Amarilyo G, La Cava A. miRNA in systemic lupus erythematosus. Clin Immunol. 2012;144:26–31. https://doi.org/10.1016/j.clim.2012.04.005.
Qu Z, Li W, Fu B. MicroRNAs in autoimmune diseases. Biomed Res Int. 2014;2014:527895. https://doi.org/10.1155/2014/527895.
Aguennouz M, Guarneri F, Oteri R, et al. Serum levels of miRNA-21-5p in vitiligo patients and effects of miRNA-21-5p on SOX5, beta-catenin, CDK2 and MITF protein expression in normal human melanocytes. J Dermatol Sci. 2021;101:22–9. https://doi.org/10.1016/j.jdermsci.2020.10.014.
Kriegsmann M, Randau TM, Gravius S, et al. Expression of miR-146a, miR-155, and miR-223 in formalin-fixed paraffin-embedded synovial tissues of patients with rheumatoid arthritis and osteoarthritis. Virchows Arch. 2016;469:93–100. https://doi.org/10.1007/s00428-016-1939-4.
Seddiki N, Brezar V, Ruffin N, et al. Role of miR-155 in the regulation of lymphocyte immune function and disease. Immunology. 2014;142:32–8. https://doi.org/10.1111/imm.12227.
Dai Y, Huang Y-S, Tang M, et al. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16:939–46. https://doi.org/10.1177/0961203307084158.
Tang Y, Luo X, Cui H, et al. MicroRNA-146a contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–75. https://doi.org/10.1002/art.24436.
Zhao X, Tang Y, Qu B, et al. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 2010;62:3425–35. https://doi.org/10.1002/art.27632.
Sonkoly E, Wei T, Janson PCJ, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS ONE. 2007;2:e610. https://doi.org/10.1371/journal.pone.0000610.
Duttagupta R, DiRienzo S, Jiang R, et al. Genome-wide maps of circulating miRNA biomarkers for ulcerative colitis. PLoS ONE. 2012;7:e31241. https://doi.org/10.1371/journal.pone.0031241.
Paraskevi A, Theodoropoulos G, Papaconstantinou I, et al. Circulating microRNA in inflammatory bowel disease. J Crohns Colitis. 2012;6:900–4. https://doi.org/10.1016/j.crohns.2012.02.006.
Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–30. https://doi.org/10.1158/0008-5472.CAN-10-2010.
Wischhusen JC, Chowdhury SM, Lee T, et al. Ultrasound-mediated delivery of miRNA-122 and anti-miRNA-21 therapeutically immunomodulates murine hepatocellular carcinoma in vivo. J Control Release. 2020;321:272–84. https://doi.org/10.1016/j.jconrel.2020.01.051.
Takagawa Y, Gen Y, Muramatsu T, et al. miR-1293, a candidate for miRNA-based cancer therapeutics, simultaneously targets BRD4 and the DNA repair pathway. Mol Ther. 2020;28:1494–505. https://doi.org/10.1016/j.ymthe.2020.04.001.
Yu J, Chen S, Niu Y, et al. Functional significance and therapeutic potential of miRNA-20b-5p in esophageal squamous cell carcinoma. Mol Ther–Nucleic Acids. 2020;21:315–31. https://doi.org/10.1016/j.omtn.2020.05.015.
Nana-Sinkam SP, Croce CM. MicroRNAs as therapeutic targets in cancer. Transl Res. 2011;157:216–25. https://doi.org/10.1016/j.trsl.2011.01.013.
Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with “antagomirs.” Nature. 2005;438:685–9. https://doi.org/10.1038/nature04303.
Cerro-Herreros E, Sabater-Arcis M, Fernandez-Costa JM, et al. miR-23b and miR-218 silencing increase muscleblind-like expression and alleviate myotonic dystrophy phenotypes in mammalian models. Nat Commun. 2018;9:2482. https://doi.org/10.1038/s41467-018-04892-4.
Thakral S, Ghoshal K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr Gene Ther. 2015;15:142–50. https://doi.org/10.2174/1566523214666141224095610.
Malik S, Lim J, Slack FJ, Braddock DT, Bahal R. Next generation miRNA inhibition using short anti-seed PNAs encapsulated in PLGA nanoparticles. J Control Release. 2020;327:406–19. https://doi.org/10.1016/j.jconrel.2020.08.026.
Fabani MM, Abreu-Goodger C, Williams D, et al. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res. 2010;38:4466–75. https://doi.org/10.1093/nar/gkq160.
Meng L, Liu C, Lü J, et al. Small RNA zippers lock miRNA molecules and block miRNA function in mammalian cells. Nat Commun. 2017;8:13964. https://doi.org/10.1038/ncomms13964.
Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat Mater. 2012;11:316–22. https://doi.org/10.1038/nmat3253.
Kota J, Chivukula RR, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. https://doi.org/10.1016/j.cell.2009.04.021.
Bader AG. miR-34 – a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. https://doi.org/10.3389/fgene.2012.00120.
Huang W. MicroRNAs: biomarkers, diagnostics, and therapeutics. Methods Mol Biol. 2017;1617:57–67. https://doi.org/10.1007/978-1-4939-7046-9_4.
Wang J, Chen J, Sen S. MicroRNA as biomarkers and diagnostics. J Cell Physiol. 2016;23:25–30. https://doi.org/10.1002/jcp.25056.
Lawrie CH. MicroRNAs in medicine. Hoboken: John Wiley & Sons Inc; 2013. https://doi.org/10.1002/9781118300312.
Palmero EI, de Campos SG, Campos M, et al. Mechanisms and role of microRNA deregulation in cancer onset and progression. Genet Mol Biol. 2011;34:363–70. https://doi.org/10.1590/S1415-47572011000300001.
Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–78. https://doi.org/10.1007/s10555-009-9188-5.
Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarkers. Mutat Res. 2011;717(1–2):85–90. https://doi.org/10.1016/j.mrfmmm.2011.03.004.
Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. https://doi.org/10.1073/pnas.0804549105.
Park NJ, Zhou H, Elashoff D, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15:5473–7. https://doi.org/10.1158/1078-0432.CCR-09-0736.
He B, Zhao Z, Cai Q, Zhang Y, et al. miRNA-based biomarkers, therapies, and resistance in cancer. Int J Biol Sci. 2020;16(14):2628–47. https://doi.org/10.7150/ijbs.47203.
Eichmüller SB, Osen W, Mandelboim O, et al. Immune modulatory microRNAs involved in tumor attack and tumor immune escape. J Natl Cancer Inst. 2017;109(10): djx034. https://doi.org/10.1093/jnci/djx034.
Yang Q, Cao W, Wang Z, et al. Regulation of cancer immune escape: the roles of miRNAs in immune checkpoint proteins. Cancer Lett. 2018;1(431):73–84. https://doi.org/10.1016/j.canlet.2018.05.015.
Harborth J, Elbashir SM, Vandenburgh K, et al. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. https://doi.org/10.1089/108729003321629638.
Landen CN Jr, Chavez-Reyes A, Bucana C, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–8. https://doi.org/10.1158/0008-5472.
Myoung SS, Kasinski AL. Strategies for safe and targeted delivery of microRNA therapeutics. In MicroRNAs in diseases and disorders 2019 May 7 (pp. 386–415). https://doi.org/10.1039/9781788016421-00386.
Cortez MA, Ivan C, Valdecanas D, et al. PDL1 regulation by p53 via miR-34. J Natl Cancer Inst. 2015;108(1):303. https://doi.org/10.1093/jnci/djv303.
Wiggins JF, Ruffino L, Kelnar K, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70(14):5923–30. https://doi.org/10.1158/0008-5472.CAN-10-0655.
Robbins M, Judge A, Ambegia E, et al. Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation. Hum Gene Ther. 2008;19(10):991–9. https://doi.org/10.1089/hum.2008.131.
Cortez MA, Anfossi S, Ramapriyan R, et al. Role of miRNAs in immune responses and immunotherapy in cancer. Genes Chromosomes Cancer. 2019;58(4):244–53. https://doi.org/10.1002/gcc.22725.
Fernandez-Piñeiro I, Badiola I, Sanchez A. Nanocarriers for microRNA delivery in cancer medicine. Biotechnol Adv. 2017;35:350–60. https://doi.org/10.1016/j.biotechadv.2017.03.002.
Vaid P, Raizada P, Saini AK, Saini RV. Biogenic silver, gold and copper nanoparticles - a sustainable green chemistry approach for cancer therapy. Sustain Chem Pharm. 2020;16:100247. https://doi.org/10.1016/j.scp.2020.100247.
Kumari R, Saini AK, Kumar A, Saini RV. Apoptosis induction in lung and prostate cancer cells through silver nanoparticles synthesized from Pinus roxburghii bioactive fraction. J Biol Inorg Chem. 2020;25:23–37. https://doi.org/10.1007/s00775-019-01729-3.
Lu C, Han HD, Mangala LS, et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18:185–97. https://doi.org/10.1016/j.ccr.2010.06.016.
Chen X, Mangala LS, Mooberry L, et al. Identifying and targeting angiogenesis-related microRNAs in ovarian cancer. Oncogene. 2019;38:6095–108. https://doi.org/10.1038/s41388-019-0862-y.
Knipe JM, Strong LE, Peppas NA. Enzyme- and pH-responsive microencapsulated nanogels for oral delivery of siRNA to induce TNF-α knockdown in the intestine. Biomacromol. 2016;17:788–97. https://doi.org/10.1021/acs.biomac.5b01518.
Liechty WB, Scheuerle RL, Vela Ramirez JE, Peppas NA. Cytoplasmic delivery of functional siRNA using pH-responsive nanoscale hydrogels. Int J Pharm. 2019;562:249–57. https://doi.org/10.1016/j.ijpharm.2019.03.013.
Forbes DC, Peppas NA. Polymeric nanocarriers for siRNA delivery to murine macrophages. Macromol Biosci. 2014;14:1096–105. https://doi.org/10.1002/mabi.201400027.
Zheng B, Chen L, Pan C-C, et al. Targeted delivery of miRNA-204-5p by PEGylated polymer nanoparticles for colon cancer therapy. Nanomedicine. 2018;13:769–85. https://doi.org/10.2217/nnm-2017-0345.
Peng Z-H, Xie Y, Wang Y, et al. Dual-function polymeric HPMA prodrugs for the delivery of miRNA. Mol Pharm. 2017;14:1395–404. https://doi.org/10.1021/acs.molpharmaceut.6b00999.
Gao S, Tian H, Guo Y, et al. miRNA oligonucleotide and sponge for miRNA-21 inhibition mediated by PEI-PLL in breast cancer therapy. Acta Biomater. 2015;25:184–93. https://doi.org/10.1016/j.actbio.2015.07.020.
Luo Q, Feng Y, Xie Y, et al. Nanoparticle–microRNA-146a-5p polyplexes ameliorate diabetic peripheral neuropathy by modulating inflammation and apoptosis. Nanomedicine. 2019;17:188–97. https://doi.org/10.1016/j.nano.2019.01.007.
Hall A, Lächelt U, Bartek J, et al. Polyplex evolution: understanding biology, optimizing performance. Mol Ther. 2017;25:1476–90. https://doi.org/10.1016/j.ymthe.2017.01.024.
Ulasov AV, Khramtsov YV, Trusov GA, et al. Properties of PEI-based polyplex nanoparticles that correlate with their transfection efficacy. Mol Ther. 2011;19:103–12. https://doi.org/10.1038/mt.2010.233.
Gong M, Liu H, Sun N, Xie Y, et al. Polyethylenimine-dextran-coated magnetic nanoparticles loaded with miR-302b suppress osteosarcoma in vitro and in vivo. Nanomedicine. 2020;15:711–23. https://doi.org/10.2217/nnm-2019-0218.
Dai Y, Zhang X. MicroRNA delivery with bioreducible polyethylenimine as a non-viral vector for breast cancer gene therapy. Macromol Biosci. 2019;19:1800445. https://doi.org/10.1002/mabi.201800445.
Mao S, Sun W, Kissel T. Chitosan-based formulations for delivery of DNA and siRNA. Adv Drug Deliv Rev. 2010;62:12–27. https://doi.org/10.1016/j.addr.2009.08.004.
Louw AM, Kolar MK, Novikova LN, et al. Chitosan polyplex mediated delivery of miRNA-124 reduces activation of microglial cells in vitro and in rat models of spinal cord injury. Nanomedicine. 2016;12:643–53. https://doi.org/10.1016/j.nano.2015.10.011.
Ning Q, Liu Y-F, Ye P-J, et al. Delivery of liver-specific miRNA-122 using a targeted macromolecular prodrug toward synergistic therapy for hepatocellular carcinoma. ACS Appl Mater Interfaces. 2019;11:10578–88. https://doi.org/10.1021/acsami.9b00634.
Gaur S, Wen Y, Song JH, et al. Chitosan nanoparticle-mediated delivery of miRNA-34a decreases prostate tumor growth in the bone and its expression induces non-canonical autophagy. Oncotarget. 2015;6:29161–77. https://doi.org/10.18632/oncotarget.4971.
Cosco D, Cilurzo F, Maiuolo J, et al. Delivery of miR-34a by chitosan/PLGA nanoplexes for the anticancer treatment of multiple myeloma. Sci Rep. 2015;5:17579. https://doi.org/10.1038/srep17579.
Arora S, Swaminathan SK, Kirtane A, et al. Synthesis, characterization, and evaluation of poly (D, L-lactide-co-glycolide)-based nanoformulation of miRNA-150: potential implications for pancreatic cancer therapy. Int J Nanomed. 2014;9:2933–42. https://doi.org/10.2147/IJN.S61949.
Wang S, Zhang J, Wang Y, Chen M. Hyaluronic acid-coated PEI-PLGA nanoparticles mediated co-delivery of doxorubicin and miR-542-3p for triple negative breast cancer therapy. Nanomedicine. 2016;12:411–20. https://doi.org/10.1016/j.nano.2015.09.014.
Cai C, Xie Y, Wu L, et al. PLGA-based dual targeted nanoparticles enhance miRNA transfection efficiency in hepatic carcinoma. Sci Rep. 2017;7:46250. https://doi.org/10.1038/srep46250.
Kapadia CH, Ioele SA, Day ES. Layer-by-layer assembled PLGA nanoparticles carrying miR-34a cargo inhibit the proliferation and cell cycle progression of triple-negative breast cancer cells. J Biomed Mater Res A. 2020;108:601–13. https://doi.org/10.1002/jbm.a.36840.
Zhang J, Chen C, Fu H, et al. MicroRNA-125a-loaded polymeric nanoparticles alleviate systemic lupus erythematosus by restoring effector/regulatory T cells balance. ACS Nano. 2020;14:4414–29. https://doi.org/10.1021/acsnano.9b09998.
Bahreyni A, Alibolandi M, Ramezani M, et al. A novel MUC1 aptamer-modified PLGA-epirubicin-PβAE-antimir-21 nanocomplex platform for targeted co-delivery of anticancer agents in vitro and in vivo. Colloids Surf B Biointerfaces. 2019;175:231–8. https://doi.org/10.1016/j.colsurfb.2018.12.006.
Sampathkumar SG, Yarema KJ. Dendrimers in cancer treatment and diagnosis. Kumar CSSR editor. Nanotechnologies for the life sciences. Wiley‐VCH Verlag GmbH & Co.; 2007. https://doi.org/10.1002/9783527610419.ntls0071.
Dzmitruk V, Apartsin E, Ihnatsyeu-Kachan A, et al. Dendrimers show promise for siRNA and microRNA therapeutics. Pharmaceutics. 2018;10:126-150. https://doi.org/10.3390/pharmaceutics10030126.
Gillies E, Frechet J. Dendrimers and dendritic polymers in drug delivery. Drug Discov Today. 2005;10:35–43. https://doi.org/10.1016/s1359-6446(04)03276-3.
Bosman AW, Janssen HM, Meijer EW. About dendrimers: structure, physical properties, and applications. Chem Rev. 1999;99:1665–88. https://doi.org/10.1021/cr970069y.
Tomalia DA, Baker H, Dewald J, et al. Dendritic macromolecules: synthesis of starburst dendrimers. Macromolecules. 1986;19:2466–8. https://doi.org/10.1021/ma00163a029.
Kim Y, Zimmerman SC. Applications of dendrimers in bio-organic chemistry. Curr Opin Chem Biol. 1998;2:733–42. https://doi.org/10.1016/s1367-5931(98)80111-7.
Smith DK, Diederich F. Functional dendrimers: unique biological mimics. Chem Eur J. 1998;4:1353–61. https://doi.org/10.1002/(sici)1521-3765(19980807)4:8%3c1353::aid-chem1353%3e3.0.co;2-0.
Stiriba S-E, Frey H, Haag R. Dendritic polymers in biomedical applications: from potential to clinical use in diagnostics and therapy. Angew Chem Int. 2002;41:1329–34. https://doi.org/10.1002/1521-3773(20020415)41:8%3c1329::aid-anie1329%3e3.0.co;2-p.
Mintzer MA, Grinstaff MW. Biomedical applications of dendrimers: a tutorial. Chem Soc Rev. 2011;40:173–90. https://doi.org/10.1039/b901839p.
Wang F, Zhang B, Zhou L, et al. Imaging dendrimer-grafted graphene oxide mediated anti-miR-21 delivery with an activatable luciferase reporter. ACS Appl Mater Interfaces. 2016;8:9014–21. https://doi.org/10.1021/acsami.6b02662.
Qian X, Ren Y, Shi Z, et al. Sequence-dependent synergistic inhibition of human glioma cell lines by combined temozolomide and miR-21 inhibitor gene therapy. Mol Pharm. 2012;9:2636–45. https://doi.org/10.1021/mp3002039.
Ren Y, Kang C-S, Yuan X-B, et al. Co-delivery of as-miR-21 and 5-FU by poly (amidoamine) dendrimer attenuates human glioma cell growth in vitro. J Biomater Sci Polym Ed. 2010;21:303–14. https://doi.org/10.1163/156856209X415828.
Abbasi E, Aval S, Akbarzadeh A, et al. Dendrimers: synthesis, applications, and properties. Nanoscale Res Lett. 2014;9:247. https://doi.org/10.1186/1556-276x-9-247.
Wu W, Hu Q, Wang M, et al. A PEGylated megamer-based microRNA delivery system activatable by stepwise microenvironment stimulation. Chem Commun. 2019;55:9363–6. https://doi.org/10.1039/c9cc03846a.
Duan Y, Xing Z, Yang J, et al. Chondroitin sulfate-functionalized polyamidoamine-mediated miR-34a delivery for inhibiting the proliferation and migration of pancreatic cancer. RSC Adv. 2016;6:70870–6. https://doi.org/10.1039/c6ra15716e.
Liu X, Li G, Su Z, et al. Poly (amido amine) is an ideal carrier of miR-7 for enhancing gene silencing effects on the EGFR pathway in U251 glioma cells. Oncol Rep. 2013;29:1387–94. https://doi.org/10.3892/or.2013.2283.
Sakurai Y, Akita H, Harashima H. Targeting tumor endothelial cells with nanoparticles. Int J Mol Sci. 2019;20:5819. https://doi.org/10.3390/ijms20235819.
Pecot CV, Calin GA, Coleman RL, et al. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11:59–67. https://doi.org/10.1038/nrc2966.
Yu HW, Cho WC. The emerging role of miRNAs in combined cancer therapy. Expert Opin Biol Ther. 2015;15:923–5. https://doi.org/10.1517/14712598.2015.1030390.
Thakur S, Saini RV, Singh P, Raizada P, et al. Nanoparticles as an emerging tool to alter the gene expression: preparation and conjugation methods. Mater Today Chem. 2020;17:100295. https://doi.org/10.1016/j.mtchem.2020.100295.
Yang C, Gao S, Song P, et al. Theranostic niosomes for efficient siRNA/microRNA delivery and activatable near-infrared fluorescent tracking of stem cells. ACS Appl Mater Interfaces. 2018;10:19494–503. https://doi.org/10.1021/acsami.8b05513.
Liang J, Zhang X, He S, et al. Sphk2 RNAi nanoparticles suppress tumor growth via downregulating cancer cell derived exosomal microRNA. J Control Release. 2018;286:348–57. https://doi.org/10.1016/j.jconrel.2018.07.039.
Piao L, Zhang M, Datta J, et al. Lipid-based nanoparticle delivery of Pre-miR-107 inhibits the tumorigenicity of head and neck squamous cell carcinoma. Mol Ther. 2012;20:1261–9. https://doi.org/10.1038/mt.2012.67.
Wu Y, Crawford M, Yu B, et al. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol Pharm. 2011;8:1381–9. https://doi.org/10.1021/mp2002076.
Hsu S-H, Yu B, Wang X, et al. Cationic lipid nanoparticles for therapeutic delivery of siRNA and miRNA to murine liver tumor. Nanomedicine. 2013;9:1169–80. https://doi.org/10.1016/j.nano.2013.05.007.
Pramanik D, Campbell NR, Karikari C, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10:1470–80. https://doi.org/10.1158/1535-7163.MCT-11-0152.
Scheideler M, Vidakovic I, Prassl R. Lipid nanocarriers for microRNA delivery. Chem Phys Lipids. 2020;226:104837. https://doi.org/10.1016/j.chemphyslip.2019.104837.
Moro M, Di Paolo D, Milione M, et al. Coated cationic lipid-nanoparticles entrapping miR-660 inhibit tumor growth in patient-derived xenografts lung cancer models. J Control Release. 2019;308:44–56. https://doi.org/10.1016/j.jconrel.2019.07.006.
Wang Z, Zhao K, Zhang Y, et al. Anti-GPC3 Antibody tagged cationic switchable lipid-based nanoparticles for the co-delivery of anti-miRNA27a and sorafenib in liver cancers. Pharm Res. 2019;36:145. https://doi.org/10.1007/s11095-019-2669-5.
Wang X, Yu B, Ren W, et al. Enhanced hepatic delivery of siRNA and microRNA using oleic acid based lipid nanoparticle formulations. J Control Release. 2013;172:690–8. https://doi.org/10.1016/j.jconrel.2013.09.027.
Acknowledgements
The authors would like to acknowledge their respective departments for their support in writing this article.
Funding
The authors are grateful to Maharishi Markandeshwar (Deemed to be University) for providing financial support for writing this review article.
Author information
Authors and Affiliations
Contributions
Conceptualization, R.V.S. and A.K.S.; writing—original draft preparation, Y.S., S.K., N.K., and R.V.S.; writing—review and editing, A.K.S., N.K., V.K.T., V.K.G., and V.S.; supervision, Y.S. and R.V.S. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sharma, Y., Saini, A.K., Kashyap, S. et al. Host miRNA and immune cell interactions: relevance in nano-therapeutics for human health. Immunol Res 70, 1–18 (2022). https://doi.org/10.1007/s12026-021-09247-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-021-09247-8